Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
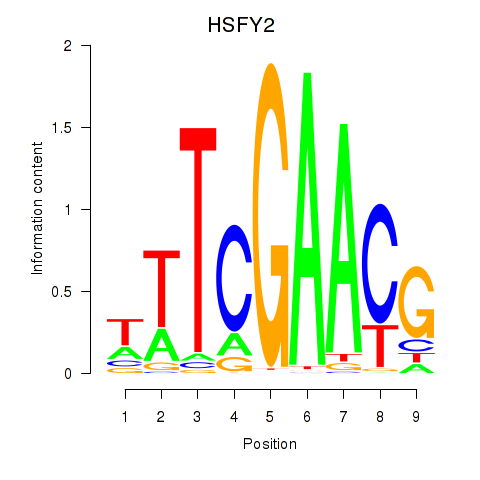
Results for HSFY2
Z-value: 0.87
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | heat shock transcription factor Y-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSFY2 | hg19_v2_chrY_-_20935572_20935621 | 0.56 | 1.3e-03 | Click! |
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_81256871 | 2.23 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr6_+_163148161 | 2.23 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr19_+_5720666 | 1.85 |
ENST00000381624.3
ENST00000381614.2 |
CATSPERD
|
catsper channel auxiliary subunit delta |
| chr7_+_105603683 | 1.68 |
ENST00000317716.9
|
CDHR3
|
cadherin-related family member 3 |
| chr10_+_70587279 | 1.56 |
ENST00000298596.6
ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1
|
storkhead box 1 |
| chr1_+_111888890 | 1.56 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr6_+_163148973 | 1.39 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr7_+_105603657 | 1.34 |
ENST00000542731.1
ENST00000343407.5 |
CDHR3
|
cadherin-related family member 3 |
| chr11_-_71318487 | 1.32 |
ENST00000343767.3
|
AP000867.1
|
AP000867.1 |
| chr6_-_10838736 | 1.28 |
ENST00000536370.1
ENST00000474039.1 |
MAK
|
male germ cell-associated kinase |
| chr9_-_39239171 | 1.26 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr1_+_111889212 | 1.21 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr4_+_52917451 | 1.19 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr1_+_1109272 | 1.13 |
ENST00000379290.1
ENST00000379289.1 |
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr7_-_6010263 | 1.09 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr19_-_55677920 | 1.08 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr2_-_209028300 | 1.07 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr7_+_89874524 | 1.07 |
ENST00000497910.1
|
C7orf63
|
chromosome 7 open reading frame 63 |
| chr6_-_10838710 | 1.04 |
ENST00000313243.2
|
MAK
|
male germ cell-associated kinase |
| chr5_+_68860949 | 1.03 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr14_-_75536182 | 1.03 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr17_+_7590734 | 1.02 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr10_+_106113515 | 1.01 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr2_-_28113965 | 1.00 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr21_-_35884573 | 0.95 |
ENST00000399286.2
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr4_-_186317034 | 0.94 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr15_+_81426588 | 0.90 |
ENST00000286732.4
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr4_-_100356291 | 0.89 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_73661364 | 0.88 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr2_+_234959376 | 0.86 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr7_-_100965011 | 0.84 |
ENST00000498704.2
ENST00000517481.1 ENST00000437644.2 ENST00000315322.4 |
RABL5
|
RAB, member RAS oncogene family-like 5 |
| chr1_-_118727781 | 0.81 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr9_-_99180597 | 0.79 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr17_-_74533963 | 0.79 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr7_+_138818490 | 0.79 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr8_-_133637624 | 0.78 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chrY_+_15016013 | 0.74 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr20_-_271304 | 0.74 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr2_-_169769787 | 0.73 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr11_+_124543694 | 0.71 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr13_-_24463530 | 0.70 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr1_-_13002348 | 0.70 |
ENST00000355096.2
|
PRAMEF6
|
PRAME family member 6 |
| chr15_+_67547163 | 0.69 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr3_-_52804872 | 0.68 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr2_+_233497931 | 0.67 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr22_-_17680472 | 0.65 |
ENST00000330232.4
|
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr8_+_81397876 | 0.64 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr11_+_61159832 | 0.63 |
ENST00000334888.5
ENST00000398979.3 |
TMEM216
|
transmembrane protein 216 |
| chr4_-_122854193 | 0.60 |
ENST00000513531.1
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr7_+_150382781 | 0.60 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr19_-_50529193 | 0.59 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr2_+_133874577 | 0.59 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr15_+_41624892 | 0.58 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr4_-_104119528 | 0.57 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr6_+_122720681 | 0.57 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr3_-_148804275 | 0.56 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr12_-_9360966 | 0.54 |
ENST00000261336.2
|
PZP
|
pregnancy-zone protein |
| chr15_+_90118723 | 0.53 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr16_+_20817839 | 0.53 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr10_+_35415851 | 0.52 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr11_+_125757556 | 0.52 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr9_+_95820966 | 0.52 |
ENST00000375472.3
ENST00000465709.1 |
SUSD3
|
sushi domain containing 3 |
| chr16_-_5147743 | 0.52 |
ENST00000587133.1
ENST00000458008.4 ENST00000427587.4 |
FAM86A
|
family with sequence similarity 86, member A |
| chr15_-_41624685 | 0.52 |
ENST00000560640.1
ENST00000220514.3 |
OIP5
|
Opa interacting protein 5 |
| chr1_-_39339777 | 0.51 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr11_+_112047087 | 0.51 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr4_+_9172135 | 0.51 |
ENST00000512047.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr7_+_102105370 | 0.51 |
ENST00000292616.5
|
LRWD1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr14_-_25519095 | 0.51 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr12_-_6961050 | 0.50 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr10_+_104614008 | 0.49 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr14_-_25519317 | 0.49 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chrX_-_24690771 | 0.48 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr4_-_75695366 | 0.48 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr11_+_102980126 | 0.48 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr1_+_61542922 | 0.48 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr6_+_127898312 | 0.48 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_+_97361218 | 0.47 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr10_+_17272608 | 0.47 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr6_+_53976285 | 0.46 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr9_-_215744 | 0.46 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr1_+_67673297 | 0.46 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr17_+_11501816 | 0.46 |
ENST00000454412.2
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr11_+_33902189 | 0.46 |
ENST00000330381.2
|
AC132216.1
|
HCG1785179; PRO1787; Uncharacterized protein |
| chr12_-_71533055 | 0.45 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr6_-_44281043 | 0.45 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr2_+_65216462 | 0.45 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr19_+_17337027 | 0.45 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr12_+_12202785 | 0.45 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr12_+_12202774 | 0.44 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr2_+_234959323 | 0.44 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr1_-_169337176 | 0.44 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr13_-_41706864 | 0.44 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr14_-_50154921 | 0.43 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr12_-_12509929 | 0.43 |
ENST00000381800.2
|
LOH12CR2
|
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr17_-_8113542 | 0.42 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr7_-_138363824 | 0.41 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr19_+_17337007 | 0.41 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr1_+_214776516 | 0.40 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr19_-_39466396 | 0.40 |
ENST00000292852.4
|
FBXO17
|
F-box protein 17 |
| chrX_-_48693955 | 0.40 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr9_+_131037623 | 0.40 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr11_-_89956227 | 0.39 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr12_-_23737534 | 0.39 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr15_+_84115868 | 0.39 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr21_-_30257669 | 0.39 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr15_+_48623600 | 0.39 |
ENST00000558813.1
ENST00000331200.3 ENST00000558472.1 |
DUT
|
deoxyuridine triphosphatase |
| chr15_+_84116106 | 0.38 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr1_-_57431679 | 0.38 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr19_-_50528584 | 0.38 |
ENST00000594092.1
ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3
|
vaccinia related kinase 3 |
| chr20_+_30327063 | 0.38 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr5_-_133706695 | 0.38 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chrX_+_36065053 | 0.37 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chr3_+_186383741 | 0.36 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr10_+_114133773 | 0.36 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr1_+_10490779 | 0.36 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr7_+_129251531 | 0.35 |
ENST00000393232.1
ENST00000353868.4 ENST00000539636.1 ENST00000454688.1 ENST00000223190.4 ENST00000311967.2 |
NRF1
|
nuclear respiratory factor 1 |
| chr6_-_29343068 | 0.35 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3 |
| chr10_+_114135004 | 0.35 |
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr3_+_184053703 | 0.35 |
ENST00000450976.1
ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A
|
family with sequence similarity 131, member A |
| chr8_+_27629459 | 0.35 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr10_+_13203543 | 0.34 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr6_-_39399087 | 0.34 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr15_+_90118685 | 0.34 |
ENST00000268138.7
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr12_+_109554386 | 0.34 |
ENST00000338432.7
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr6_+_87646995 | 0.33 |
ENST00000305344.5
|
HTR1E
|
5-hydroxytryptamine (serotonin) receptor 1E, G protein-coupled |
| chr1_+_15573757 | 0.33 |
ENST00000358897.4
ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr3_+_2933893 | 0.33 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr22_-_36013368 | 0.33 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr16_+_15596123 | 0.33 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_+_184826418 | 0.33 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr11_+_58910295 | 0.32 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr20_+_30640004 | 0.32 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr1_-_197115818 | 0.32 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr7_-_154863264 | 0.32 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr2_+_124782857 | 0.31 |
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5 |
| chr1_-_150207017 | 0.31 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_-_100356551 | 0.31 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_14956506 | 0.31 |
ENST00000330828.2
|
C12orf60
|
chromosome 12 open reading frame 60 |
| chr10_+_118350522 | 0.31 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr15_+_27112058 | 0.31 |
ENST00000355395.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr19_+_58193388 | 0.31 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr4_-_100356844 | 0.31 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr16_+_81040103 | 0.30 |
ENST00000305850.5
ENST00000299572.5 |
CENPN
|
centromere protein N |
| chr15_+_67547113 | 0.30 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr11_+_102980251 | 0.30 |
ENST00000334267.7
ENST00000398093.3 |
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr11_-_108464321 | 0.30 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr15_-_45670924 | 0.30 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_+_133918175 | 0.29 |
ENST00000298622.4
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr20_+_10199468 | 0.29 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr16_-_20416053 | 0.29 |
ENST00000302451.4
|
PDILT
|
protein disulfide isomerase-like, testis expressed |
| chr2_+_201754050 | 0.29 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr20_+_30639991 | 0.28 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr17_-_38574169 | 0.28 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr16_+_2479390 | 0.28 |
ENST00000397066.4
|
CCNF
|
cyclin F |
| chr1_+_160765860 | 0.28 |
ENST00000368037.5
|
LY9
|
lymphocyte antigen 9 |
| chr10_+_7745232 | 0.28 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_+_65479702 | 0.28 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr12_+_123459127 | 0.28 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr8_+_95835438 | 0.27 |
ENST00000521860.1
ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8
|
integrator complex subunit 8 |
| chr7_-_102789629 | 0.27 |
ENST00000417955.1
ENST00000341533.4 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr10_+_35416223 | 0.27 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr22_-_41985865 | 0.27 |
ENST00000216259.7
|
PMM1
|
phosphomannomutase 1 |
| chr10_-_43133950 | 0.27 |
ENST00000359467.3
|
ZNF33B
|
zinc finger protein 33B |
| chr14_+_21525981 | 0.27 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr18_-_53253323 | 0.27 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr12_-_88535747 | 0.27 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr1_-_149900122 | 0.27 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr15_+_40886199 | 0.27 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr4_-_105416039 | 0.26 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr16_-_67969888 | 0.26 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr10_+_105881906 | 0.26 |
ENST00000369727.3
ENST00000336358.5 |
SFR1
|
SWI5-dependent recombination repair 1 |
| chr16_-_18462221 | 0.25 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr18_+_61637159 | 0.25 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr7_+_120590803 | 0.25 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr8_+_42552533 | 0.25 |
ENST00000289957.2
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr1_-_26232522 | 0.25 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr11_+_71498552 | 0.25 |
ENST00000346333.6
ENST00000359244.4 ENST00000426628.2 |
FAM86C1
|
family with sequence similarity 86, member C1 |
| chr3_-_72149553 | 0.24 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr11_+_43702322 | 0.24 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr2_-_136633940 | 0.24 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr9_+_71939488 | 0.24 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr6_+_7389694 | 0.24 |
ENST00000379834.2
|
RIOK1
|
RIO kinase 1 |
| chr17_+_30771279 | 0.23 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr3_-_146262352 | 0.23 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr4_-_1166954 | 0.23 |
ENST00000514490.1
ENST00000431380.1 ENST00000503765.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr1_+_28052456 | 0.23 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr1_-_226496772 | 0.23 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr10_+_105881779 | 0.23 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr3_-_133614597 | 0.23 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr1_-_169863016 | 0.22 |
ENST00000367772.4
ENST00000367771.6 |
SCYL3
|
SCY1-like 3 (S. cerevisiae) |
| chr9_+_99212403 | 0.22 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr4_-_168155169 | 0.22 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_-_7679633 | 0.22 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr14_+_96722539 | 0.22 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr3_-_146262365 | 0.22 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr8_+_80523962 | 0.22 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr5_-_37371278 | 0.22 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr3_-_149293990 | 0.22 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chrX_+_46940254 | 0.22 |
ENST00000336169.3
|
RGN
|
regucalcin |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 1.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.3 | 1.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 2.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 1.6 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.2 | 0.6 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 1.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 1.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 0.5 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.2 | 1.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.5 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.8 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.4 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.7 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.6 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.5 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.5 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.1 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.2 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.3 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.9 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.4 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.0 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.7 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.2 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 4.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.5 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0048378 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.2 | GO:0051198 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 1.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 0.9 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.2 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 3.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.5 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 4.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 2.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.6 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 0.8 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.2 | 1.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.2 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 6.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.5 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.5 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 2.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |