Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
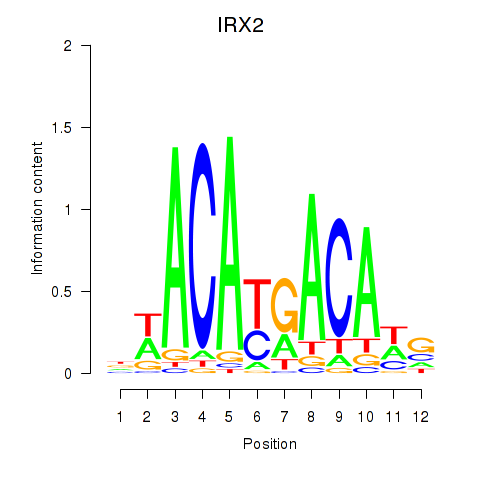
Results for IRX2
Z-value: 0.43
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.8 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg19_v2_chr5_-_2751762_2751784 | -0.11 | 5.5e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_112565703 | 1.10 |
ENST00000488794.1
|
CD200R1L
|
CD200 receptor 1-like |
| chr16_+_82068830 | 1.02 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr10_-_28270795 | 0.87 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr22_-_36013368 | 0.87 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr5_+_36608422 | 0.84 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_+_88117683 | 0.81 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr19_+_13134772 | 0.78 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr13_+_35516390 | 0.78 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr14_+_75536280 | 0.74 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_+_75536335 | 0.68 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr4_-_70505358 | 0.63 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chrX_-_117119243 | 0.62 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr17_+_7761301 | 0.61 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr10_+_134150835 | 0.61 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr3_-_151102529 | 0.58 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr4_+_69681710 | 0.56 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr11_+_103907308 | 0.56 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr17_+_7761013 | 0.56 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr5_+_156696362 | 0.55 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr2_+_159651821 | 0.53 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr13_-_46679185 | 0.53 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 0.52 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr4_-_38806404 | 0.48 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr10_+_127661942 | 0.47 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr10_-_100027943 | 0.45 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr8_+_94767072 | 0.41 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr17_+_56232494 | 0.40 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr10_+_70320413 | 0.40 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr3_-_113160334 | 0.37 |
ENST00000393845.2
ENST00000295868.2 |
WDR52
|
WD repeat domain 52 |
| chr7_-_105332084 | 0.36 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr15_-_45670924 | 0.36 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr12_-_112450915 | 0.35 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr3_-_167371740 | 0.35 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr4_+_83821835 | 0.34 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr6_-_53013620 | 0.33 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr11_+_6866883 | 0.33 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr5_+_147258266 | 0.30 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chrX_-_21676442 | 0.30 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr3_-_114343768 | 0.29 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_54405773 | 0.28 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_207095324 | 0.27 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_-_92371839 | 0.27 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr2_+_133874577 | 0.26 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr4_-_168155417 | 0.26 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr18_-_53070913 | 0.25 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr3_+_108541545 | 0.25 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_130598059 | 0.25 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chrX_+_9431324 | 0.24 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr8_+_81397876 | 0.24 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr8_+_24151553 | 0.23 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_+_166958346 | 0.23 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_-_151148492 | 0.23 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr2_-_233877912 | 0.23 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr10_+_46997926 | 0.22 |
ENST00000374314.4
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr22_+_23046750 | 0.22 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr10_+_96522361 | 0.22 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr17_-_26127525 | 0.22 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr1_+_166958497 | 0.22 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_-_151148442 | 0.21 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr15_-_20193370 | 0.21 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr4_+_189060573 | 0.20 |
ENST00000332517.3
|
TRIML1
|
tripartite motif family-like 1 |
| chr16_-_4401258 | 0.20 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr22_+_29168652 | 0.19 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr7_+_150065278 | 0.17 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr3_-_186262166 | 0.17 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr14_+_62462541 | 0.17 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr12_+_40787194 | 0.16 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr14_+_50999744 | 0.16 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr1_-_207095212 | 0.16 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr6_+_30908747 | 0.15 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr4_+_88896819 | 0.14 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr11_+_98891797 | 0.14 |
ENST00000527185.1
ENST00000528682.1 ENST00000524871.1 |
CNTN5
|
contactin 5 |
| chr19_-_35719609 | 0.14 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr3_-_194072019 | 0.14 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chrX_-_24045303 | 0.13 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr11_-_110561721 | 0.12 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr17_+_44590076 | 0.12 |
ENST00000333412.3
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr9_-_13175823 | 0.12 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chrX_+_150869023 | 0.11 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chrX_-_118739835 | 0.11 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr10_+_90660832 | 0.11 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr14_-_75536182 | 0.11 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chrX_+_148855726 | 0.11 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr19_-_38720294 | 0.11 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr3_-_58419537 | 0.10 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr5_-_94417314 | 0.10 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr3_-_11645925 | 0.10 |
ENST00000413604.1
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chrX_-_148676974 | 0.10 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr1_-_157811588 | 0.09 |
ENST00000368174.4
|
CD5L
|
CD5 molecule-like |
| chr4_-_153332886 | 0.09 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr7_-_107770794 | 0.09 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr15_-_44486632 | 0.09 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr17_-_27054952 | 0.09 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chr11_+_77532233 | 0.09 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr15_+_44580955 | 0.09 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr14_+_52327109 | 0.09 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_-_3504587 | 0.08 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr3_+_132843652 | 0.08 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr17_-_2614927 | 0.08 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr18_+_5748793 | 0.08 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr1_+_206317450 | 0.08 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr1_-_238649319 | 0.07 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr6_-_27841289 | 0.07 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr2_-_46844242 | 0.07 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr5_+_167913450 | 0.07 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chrX_-_100129320 | 0.07 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr14_+_23790655 | 0.07 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr1_+_204839959 | 0.06 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr19_-_38720354 | 0.06 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr4_+_119606523 | 0.06 |
ENST00000388822.5
ENST00000506780.1 ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr6_+_13272904 | 0.06 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_+_66218212 | 0.06 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr2_-_98280383 | 0.06 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr12_+_32655110 | 0.06 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_-_33140498 | 0.06 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr2_-_46844159 | 0.06 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr10_+_102672712 | 0.05 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr9_-_35812236 | 0.05 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr1_+_215747118 | 0.05 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr12_-_112856623 | 0.05 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr11_-_118305921 | 0.04 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr2_-_70520539 | 0.04 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr10_+_1095416 | 0.04 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr8_-_124428569 | 0.04 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr2_-_109605663 | 0.04 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr9_+_35792151 | 0.04 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr17_+_37856299 | 0.04 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr1_+_67673297 | 0.04 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr4_+_159131630 | 0.04 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr20_+_56136136 | 0.04 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chrX_+_19373700 | 0.04 |
ENST00000379804.1
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr4_-_80247162 | 0.03 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr8_-_86253888 | 0.03 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr3_+_31574189 | 0.03 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr8_+_40018977 | 0.03 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr8_+_67104323 | 0.03 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr16_+_69373323 | 0.03 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr8_+_81397846 | 0.03 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr5_-_39364586 | 0.03 |
ENST00000263408.4
|
C9
|
complement component 9 |
| chr18_+_50278430 | 0.02 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr2_+_46844290 | 0.02 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr13_+_28519343 | 0.02 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr12_+_53773944 | 0.02 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr17_+_74075263 | 0.02 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr20_-_31592113 | 0.02 |
ENST00000420875.1
ENST00000375519.2 ENST00000375523.3 ENST00000356173.3 |
SUN5
|
Sad1 and UNC84 domain containing 5 |
| chr12_-_91505608 | 0.02 |
ENST00000266718.4
|
LUM
|
lumican |
| chr1_+_29138654 | 0.02 |
ENST00000234961.2
|
OPRD1
|
opioid receptor, delta 1 |
| chr16_+_21244986 | 0.01 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr1_+_113010056 | 0.01 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr19_-_29704448 | 0.01 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr9_-_113100088 | 0.01 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr4_-_83821676 | 0.01 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr17_-_5322786 | 0.00 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr1_+_152691998 | 0.00 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr19_+_3762645 | 0.00 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr4_+_187187337 | 0.00 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr3_+_46616017 | 0.00 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr18_+_55018044 | 0.00 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_138721850 | 0.00 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.5 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.6 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.9 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.4 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.3 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |