Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
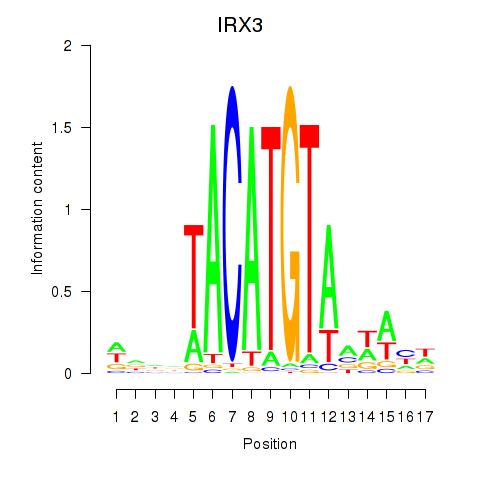
Results for IRX3
Z-value: 0.83
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.07 | 7.2e-01 | Click! |
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_69681710 | 2.40 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_-_89545079 | 1.59 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr4_+_70861647 | 1.48 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr19_-_9092018 | 1.38 |
ENST00000397910.4
|
MUC16
|
mucin 16, cell surface associated |
| chr12_-_91573132 | 1.33 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr13_-_20080080 | 1.29 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr10_+_118305435 | 1.29 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr4_+_69962185 | 1.28 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr10_+_118187379 | 1.27 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr12_-_91574142 | 1.22 |
ENST00000547937.1
|
DCN
|
decorin |
| chr12_-_10282836 | 1.17 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_-_70725856 | 1.16 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr9_+_105757590 | 1.16 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr19_-_22379753 | 1.15 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr2_+_89975669 | 1.15 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr4_-_70080449 | 1.14 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chrX_-_15620192 | 1.07 |
ENST00000427411.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr13_+_25254545 | 1.05 |
ENST00000218548.6
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr4_+_70146217 | 1.04 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr12_-_10282681 | 1.03 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_-_10282742 | 0.97 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr16_+_19421803 | 0.94 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr6_-_52705641 | 0.93 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr13_+_25254693 | 0.91 |
ENST00000381946.3
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr5_+_140235469 | 0.91 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr12_+_21207503 | 0.85 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_+_95326416 | 0.84 |
ENST00000371481.4
ENST00000371483.4 ENST00000604414.1 |
FFAR4
|
free fatty acid receptor 4 |
| chr12_-_91573316 | 0.84 |
ENST00000393155.1
|
DCN
|
decorin |
| chr4_+_69962212 | 0.82 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr9_+_90112767 | 0.79 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr7_-_99573640 | 0.77 |
ENST00000411734.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr12_+_69742121 | 0.76 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr5_+_147443534 | 0.75 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr16_-_20709066 | 0.70 |
ENST00000520010.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr7_-_99573677 | 0.70 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_-_31902614 | 0.68 |
ENST00000596131.1
|
AC114494.1
|
HCG1787699; Uncharacterized protein |
| chr11_+_27062502 | 0.68 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr11_-_111782484 | 0.67 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr11_+_27076764 | 0.67 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_-_106453155 | 0.67 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr5_+_140220769 | 0.66 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr11_-_111782696 | 0.65 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr22_-_18923655 | 0.64 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr4_-_170924888 | 0.63 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_+_50691437 | 0.63 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr8_+_84824920 | 0.60 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr7_+_13141097 | 0.60 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr12_-_127174806 | 0.58 |
ENST00000545853.1
ENST00000537478.1 |
RP11-407A16.3
|
RP11-407A16.3 |
| chr2_-_16804320 | 0.58 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr12_-_71182695 | 0.57 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_+_34639793 | 0.57 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr6_+_29407083 | 0.56 |
ENST00000444197.2
|
OR10C1
|
olfactory receptor, family 10, subfamily C, member 1 (gene/pseudogene) |
| chr3_-_127541194 | 0.55 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr12_-_10605929 | 0.55 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_+_41361616 | 0.55 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_+_80267973 | 0.54 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr21_-_43528548 | 0.53 |
ENST00000329015.2
|
C21orf128
|
chromosome 21 open reading frame 128 |
| chr6_-_47009996 | 0.53 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr11_+_57365150 | 0.52 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr9_-_21975088 | 0.52 |
ENST00000304494.5
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr8_+_120885949 | 0.52 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr20_-_13971255 | 0.52 |
ENST00000284951.5
ENST00000378072.5 |
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr12_-_13248598 | 0.51 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr2_+_106679690 | 0.51 |
ENST00000409944.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr12_-_91546926 | 0.49 |
ENST00000550758.1
|
DCN
|
decorin |
| chr11_-_63330842 | 0.49 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr9_-_37465396 | 0.49 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr22_+_23161491 | 0.49 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr20_+_2276639 | 0.49 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr13_-_46716969 | 0.49 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_+_29542557 | 0.49 |
ENST00000550906.1
|
OVCH1-AS1
|
OVCH1 antisense RNA 1 |
| chr18_+_9885961 | 0.48 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr3_-_185270383 | 0.48 |
ENST00000296252.4
|
LIPH
|
lipase, member H |
| chr2_-_88125471 | 0.48 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr19_-_44388116 | 0.47 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr21_-_33975547 | 0.47 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr10_+_82009466 | 0.46 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr1_-_46598284 | 0.46 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr14_-_106926724 | 0.45 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr1_-_46598371 | 0.45 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr4_-_103998439 | 0.45 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_-_9683238 | 0.45 |
ENST00000571771.1
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr1_-_152086556 | 0.44 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr16_+_20775358 | 0.43 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr13_+_97928395 | 0.43 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr19_+_53761545 | 0.42 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr22_-_36556821 | 0.42 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr13_+_113030625 | 0.42 |
ENST00000283550.3
|
SPACA7
|
sperm acrosome associated 7 |
| chr19_-_10491130 | 0.41 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr4_-_103997862 | 0.41 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr6_-_35765088 | 0.41 |
ENST00000259938.2
|
CLPS
|
colipase, pancreatic |
| chr22_+_51176624 | 0.41 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr7_-_92777606 | 0.40 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr7_-_16921601 | 0.40 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr1_+_220863187 | 0.39 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr11_-_49230184 | 0.39 |
ENST00000340334.7
ENST00000256999.2 |
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chrX_-_77150911 | 0.39 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr19_-_54974894 | 0.39 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr20_-_18477862 | 0.38 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr11_-_27384737 | 0.38 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr10_-_101841588 | 0.38 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr1_+_145524891 | 0.38 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr11_+_60048053 | 0.37 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr4_-_84035905 | 0.37 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr6_+_131958436 | 0.36 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr8_+_9009296 | 0.36 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr13_-_38172863 | 0.36 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr5_+_175490540 | 0.36 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr3_+_51895621 | 0.36 |
ENST00000333127.3
|
IQCF2
|
IQ motif containing F2 |
| chr11_-_26588634 | 0.35 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr3_-_185270342 | 0.35 |
ENST00000424591.2
|
LIPH
|
lipase, member H |
| chr9_+_130026756 | 0.34 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_127441406 | 0.34 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr15_-_56757329 | 0.34 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr14_+_22636283 | 0.34 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chr12_+_56114189 | 0.34 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr21_-_43816052 | 0.33 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr12_+_20963632 | 0.33 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_+_41540160 | 0.33 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_+_94727048 | 0.33 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr8_+_9009202 | 0.32 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr8_+_52730143 | 0.32 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr3_-_151102529 | 0.32 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr9_+_99690592 | 0.32 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr2_+_101591314 | 0.31 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr10_-_96829246 | 0.31 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr16_+_56995854 | 0.31 |
ENST00000566128.1
|
CETP
|
cholesteryl ester transfer protein, plasma |
| chr15_-_81616446 | 0.31 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr6_-_32920794 | 0.31 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr12_-_91348949 | 0.31 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr17_-_34207295 | 0.31 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr3_-_151047327 | 0.30 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr16_-_72206034 | 0.30 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr12_+_56114151 | 0.30 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr14_-_106963409 | 0.29 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr10_+_96698406 | 0.29 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr10_+_91092241 | 0.29 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr3_+_53528659 | 0.29 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr2_-_8464760 | 0.29 |
ENST00000430192.1
|
LINC00299
|
long intergenic non-protein coding RNA 299 |
| chr7_-_112430427 | 0.29 |
ENST00000449743.1
ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168
|
transmembrane protein 168 |
| chr1_+_202317815 | 0.29 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_+_78056412 | 0.29 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr8_-_16043780 | 0.28 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr1_-_89488510 | 0.28 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr3_+_8543533 | 0.28 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr8_+_12803176 | 0.28 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr19_-_40596828 | 0.28 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr5_-_131879205 | 0.28 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr4_-_103998060 | 0.28 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr1_+_160709076 | 0.28 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr2_-_233415220 | 0.28 |
ENST00000408957.3
|
TIGD1
|
tigger transposable element derived 1 |
| chr12_+_8309630 | 0.27 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr14_-_23624511 | 0.27 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr7_-_111202511 | 0.27 |
ENST00000452895.1
ENST00000452753.1 ENST00000331762.3 |
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_-_161337662 | 0.27 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr1_+_160709055 | 0.27 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr7_+_37960163 | 0.27 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr2_-_158300556 | 0.27 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr5_-_177207634 | 0.27 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr8_+_7801144 | 0.27 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr1_-_112032284 | 0.27 |
ENST00000414219.1
|
ADORA3
|
adenosine A3 receptor |
| chr17_-_26127525 | 0.26 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr2_-_228497888 | 0.26 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr9_-_136203235 | 0.26 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr16_-_67224002 | 0.26 |
ENST00000563889.1
ENST00000564418.1 ENST00000545725.2 ENST00000314586.6 |
EXOC3L1
|
exocyst complex component 3-like 1 |
| chr3_+_142838091 | 0.26 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr9_-_97356075 | 0.26 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr17_+_56232494 | 0.26 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr13_+_76362974 | 0.25 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr14_-_106471723 | 0.25 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr1_-_213020991 | 0.25 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr19_+_55281260 | 0.25 |
ENST00000336077.6
ENST00000291633.7 |
KIR2DL1
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1 |
| chr10_+_29577974 | 0.25 |
ENST00000375500.3
|
LYZL1
|
lysozyme-like 1 |
| chr13_+_21714653 | 0.25 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr3_-_74570291 | 0.25 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr1_+_87012753 | 0.25 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr15_-_22473353 | 0.24 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr5_-_146302078 | 0.24 |
ENST00000508545.2
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_234545148 | 0.24 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr12_+_10365404 | 0.24 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_55248289 | 0.24 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr14_+_100485712 | 0.24 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr17_-_61905005 | 0.24 |
ENST00000584574.1
ENST00000585145.1 ENST00000427159.2 |
FTSJ3
|
FtsJ homolog 3 (E. coli) |
| chr16_-_3493528 | 0.23 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr1_+_26644441 | 0.23 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr1_+_160709029 | 0.23 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr14_-_51562745 | 0.23 |
ENST00000298355.3
|
TRIM9
|
tripartite motif containing 9 |
| chr1_+_15668240 | 0.23 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr8_+_24151620 | 0.22 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_-_147211226 | 0.22 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr1_-_227505289 | 0.22 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr14_-_71001708 | 0.22 |
ENST00000256389.3
|
ADAM20
|
ADAM metallopeptidase domain 20 |
| chrX_+_65382381 | 0.22 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr2_-_231090344 | 0.22 |
ENST00000540870.1
ENST00000416610.1 |
SP110
|
SP110 nuclear body protein |
| chr1_-_112106578 | 0.22 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr20_+_54933971 | 0.21 |
ENST00000371384.3
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210, member B |
| chr11_-_65150103 | 0.21 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr1_+_202431859 | 0.21 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr17_-_42327236 | 0.21 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr2_+_217735493 | 0.21 |
ENST00000456163.1
|
AC007557.1
|
HCG1816075; Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 3.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.7 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 1.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 0.5 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 3.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 1.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.6 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 1.5 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 2.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:1900425 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.1 | 2.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.6 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.5 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.3 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.6 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.5 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 4.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.9 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.3 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 2.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0009743 | response to carbohydrate(GO:0009743) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0030593 | neutrophil chemotaxis(GO:0030593) granulocyte chemotaxis(GO:0071621) |
| 0.0 | 0.1 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.0 | 0.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.0 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.4 | 1.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.3 | 4.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.5 | 2.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 1.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.3 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.3 | 1.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.2 | 7.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 3.9 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.4 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.4 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 2.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 3.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 1.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 2.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 2.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |