Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
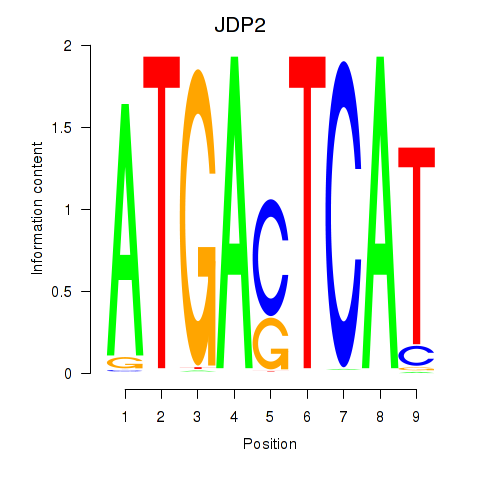
Results for JDP2
Z-value: 0.67
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_15016725 | 3.11 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr11_-_102668879 | 1.83 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr10_+_17270214 | 1.52 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr12_+_7023735 | 1.36 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_+_7023491 | 1.35 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr9_-_124991124 | 1.19 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr15_+_89182178 | 1.03 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr13_+_32838801 | 0.96 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr15_+_89181974 | 0.84 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_+_127661942 | 0.83 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr20_-_17539456 | 0.79 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr1_-_204121298 | 0.78 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204121102 | 0.78 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204121013 | 0.77 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr6_+_33043703 | 0.76 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr16_+_15596123 | 0.75 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr9_-_123638633 | 0.74 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr7_-_150721570 | 0.69 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr3_-_45957088 | 0.67 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr9_-_123639304 | 0.66 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr6_-_161695074 | 0.65 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr8_-_27472198 | 0.64 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr6_-_161695042 | 0.62 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_35992780 | 0.61 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr15_+_89182156 | 0.57 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_-_82708519 | 0.57 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr6_-_119031228 | 0.53 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr19_-_36019123 | 0.53 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_-_153521597 | 0.52 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr12_-_54813229 | 0.51 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr3_-_45957534 | 0.51 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr17_+_4853442 | 0.50 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_10262442 | 0.48 |
ENST00000360566.2
|
RRM2
|
ribonucleotide reductase M2 |
| chr3_-_18466787 | 0.48 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr12_-_75784669 | 0.47 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr1_-_153521714 | 0.46 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr4_-_987217 | 0.43 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_176237478 | 0.42 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr10_+_1102303 | 0.39 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr3_+_124223586 | 0.38 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chrX_-_20074895 | 0.36 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr7_-_80551671 | 0.35 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr19_-_45927622 | 0.33 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr11_+_5009424 | 0.32 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr10_+_52152766 | 0.32 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr11_-_102826434 | 0.32 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr3_-_18466026 | 0.31 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr4_+_71588372 | 0.31 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr16_+_3692931 | 0.31 |
ENST00000407479.1
|
DNASE1
|
deoxyribonuclease I |
| chr17_-_27045427 | 0.30 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr16_+_31366455 | 0.30 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr17_+_66245341 | 0.29 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr11_+_82783097 | 0.29 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr3_-_195997410 | 0.29 |
ENST00000419333.1
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr17_-_27044903 | 0.29 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr12_-_719573 | 0.29 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr8_-_42623747 | 0.28 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr17_-_27045165 | 0.27 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr17_-_27044810 | 0.27 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr17_-_27045405 | 0.26 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr5_+_73109339 | 0.26 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr8_-_42623924 | 0.26 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_-_244006528 | 0.26 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr8_+_70404996 | 0.26 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr16_+_31366536 | 0.25 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr16_-_28518153 | 0.25 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr11_-_82782861 | 0.25 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr7_-_122342966 | 0.24 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr2_+_28615669 | 0.23 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr7_-_122342988 | 0.23 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chrX_-_15288154 | 0.23 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr6_+_72596406 | 0.23 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_65593784 | 0.22 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr8_+_15397732 | 0.21 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr1_-_246580705 | 0.21 |
ENST00000541742.1
|
SMYD3
|
SET and MYND domain containing 3 |
| chr2_-_145275211 | 0.21 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_131200810 | 0.21 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr20_+_30946106 | 0.21 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr16_+_30077055 | 0.21 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_50300427 | 0.20 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr1_+_27719148 | 0.20 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr4_-_146019693 | 0.20 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr1_+_213224572 | 0.20 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr3_+_57094469 | 0.19 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr7_-_55606346 | 0.18 |
ENST00000545390.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr9_-_91793675 | 0.18 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr8_-_141774467 | 0.18 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr18_-_74207146 | 0.18 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr6_-_56819385 | 0.17 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr9_-_74979420 | 0.17 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr11_-_57417405 | 0.17 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr4_-_152147579 | 0.16 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr18_+_21693306 | 0.16 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr3_-_48632593 | 0.16 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_-_102714534 | 0.16 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr19_-_51538148 | 0.16 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr12_-_53343602 | 0.16 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr3_+_69134124 | 0.16 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr2_-_220173685 | 0.16 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr14_-_74226961 | 0.16 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr7_+_872107 | 0.16 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr13_-_41593425 | 0.15 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr7_+_97361388 | 0.15 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr16_+_30077098 | 0.15 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr6_+_122720681 | 0.15 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr7_+_134528635 | 0.14 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr12_-_6960407 | 0.14 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr9_-_34662651 | 0.14 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr11_-_2924720 | 0.14 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_+_74757050 | 0.14 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr7_+_28725585 | 0.14 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_+_13359819 | 0.14 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr1_-_209824643 | 0.14 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr1_-_13007420 | 0.14 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr11_-_68780824 | 0.14 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr17_-_28257080 | 0.13 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr10_-_4285923 | 0.13 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr11_+_20620946 | 0.13 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr19_-_51538118 | 0.13 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr1_-_13117736 | 0.13 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr11_-_119599794 | 0.12 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr16_-_31161380 | 0.12 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr17_-_39296739 | 0.12 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr5_+_35856951 | 0.12 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr5_+_135394840 | 0.12 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr10_+_121410882 | 0.12 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr19_-_46285646 | 0.12 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr18_+_61442629 | 0.11 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr5_-_75919253 | 0.11 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr3_-_98241358 | 0.11 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr7_+_97361218 | 0.11 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr3_-_98241760 | 0.11 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr8_-_110988070 | 0.11 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr5_+_177540444 | 0.11 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr3_+_191046810 | 0.10 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr15_+_22892663 | 0.10 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr10_+_1102721 | 0.10 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr12_+_6644443 | 0.10 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_+_75446819 | 0.10 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr3_+_69134080 | 0.10 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr7_+_48128194 | 0.09 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr4_+_84457250 | 0.09 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_15943995 | 0.09 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr6_+_56819773 | 0.09 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr17_+_35851570 | 0.09 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr4_-_36246060 | 0.09 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_49755019 | 0.09 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr4_-_100356291 | 0.09 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_+_56820018 | 0.09 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr17_-_1553346 | 0.09 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr6_-_31704282 | 0.08 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr17_-_8263538 | 0.08 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr9_+_140135665 | 0.08 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr20_+_42136308 | 0.08 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr1_+_151227179 | 0.08 |
ENST00000368884.3
ENST00000368881.4 |
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr7_+_48128316 | 0.08 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr12_+_100867694 | 0.08 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr22_+_20861858 | 0.08 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr3_-_48601206 | 0.08 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr5_+_72143988 | 0.08 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_+_101983176 | 0.08 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr11_-_124180733 | 0.07 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1 |
| chr17_+_25799008 | 0.07 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr6_+_292051 | 0.07 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr3_-_32022733 | 0.07 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chrX_+_57618269 | 0.07 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr16_+_89988259 | 0.06 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr4_+_84457529 | 0.06 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_156084461 | 0.06 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr4_-_122085469 | 0.06 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_156096336 | 0.06 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr12_-_57914275 | 0.05 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr5_+_145317356 | 0.05 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr10_+_88428206 | 0.05 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr17_-_53800217 | 0.05 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr3_+_48507621 | 0.05 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr1_-_51796987 | 0.05 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr2_+_47596287 | 0.05 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr22_+_39916558 | 0.05 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr12_+_52695617 | 0.05 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr11_-_62323702 | 0.04 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr2_-_175712270 | 0.04 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr3_-_151034734 | 0.04 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chrX_+_133371077 | 0.03 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr4_+_86525299 | 0.03 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr21_+_26934165 | 0.03 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr17_+_74381343 | 0.03 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr1_-_108231101 | 0.03 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr12_-_48152611 | 0.03 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr3_+_48507210 | 0.03 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr11_-_66103932 | 0.03 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_111148241 | 0.03 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr6_-_84140757 | 0.03 |
ENST00000541327.1
ENST00000369705.3 ENST00000543031.1 |
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr17_+_79650962 | 0.03 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr3_-_114343768 | 0.03 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_-_76817036 | 0.03 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr22_-_39715600 | 0.02 |
ENST00000427905.1
ENST00000402527.1 ENST00000216146.4 |
RPL3
|
ribosomal protein L3 |
| chr11_-_2924970 | 0.02 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr19_+_3366547 | 0.02 |
ENST00000341919.3
ENST00000590282.1 ENST00000443272.2 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr2_+_202122703 | 0.02 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 2.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.6 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.4 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 2.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.7 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 2.0 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 2.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0097240 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0071500 | negative regulation of CREB transcription factor activity(GO:0032792) cellular response to nitrosative stress(GO:0071500) regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 3.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 1.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 1.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |