Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
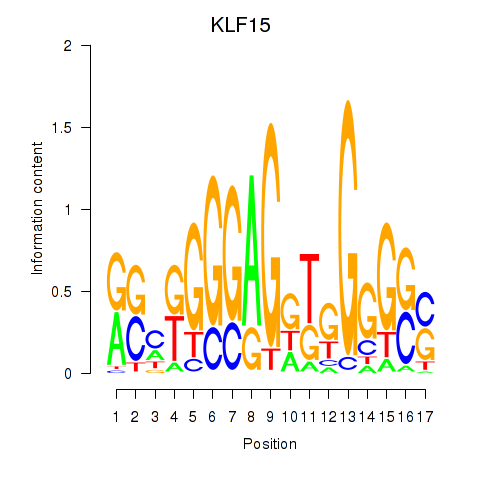
Results for KLF15
Z-value: 0.61
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | Kruppel like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | -0.02 | 9.2e-01 | Click! |
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29595779 | 2.77 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_+_1874200 | 1.33 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
| chr7_+_94285637 | 1.19 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr14_-_65439132 | 1.15 |
ENST00000533601.2
|
RAB15
|
RAB15, member RAS oncogene family |
| chr7_-_154863264 | 1.06 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr22_-_50970506 | 0.99 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr4_+_74702214 | 0.98 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr8_+_81397876 | 0.92 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr18_-_45935663 | 0.89 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr3_-_12200851 | 0.87 |
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr18_-_74207146 | 0.85 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chrX_-_48693955 | 0.80 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr3_+_238273 | 0.79 |
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr14_-_65438865 | 0.78 |
ENST00000267512.5
|
RAB15
|
RAB15, member RAS oncogene family |
| chr7_-_99869799 | 0.75 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr7_-_94285511 | 0.73 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr4_+_55524085 | 0.71 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr19_+_11457232 | 0.70 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr15_-_45480153 | 0.66 |
ENST00000560471.1
ENST00000560540.1 |
SHF
|
Src homology 2 domain containing F |
| chr7_-_94285402 | 0.66 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr6_-_80247105 | 0.65 |
ENST00000369846.4
|
LCA5
|
Leber congenital amaurosis 5 |
| chr17_-_4545170 | 0.64 |
ENST00000576394.1
ENST00000574640.1 |
ALOX15
|
arachidonate 15-lipoxygenase |
| chr6_-_80247140 | 0.62 |
ENST00000392959.1
ENST00000467898.3 |
LCA5
|
Leber congenital amaurosis 5 |
| chr7_-_94285472 | 0.60 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr12_+_19282643 | 0.60 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_-_76256557 | 0.58 |
ENST00000275569.4
ENST00000310842.4 |
POMZP3
|
POM121 and ZP3 fusion |
| chr17_+_55333876 | 0.52 |
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2 |
| chr1_-_47407097 | 0.51 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr11_+_45918092 | 0.51 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr17_-_41984835 | 0.50 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr10_+_35415851 | 0.48 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr17_+_30593195 | 0.48 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr5_+_17217669 | 0.47 |
ENST00000322611.3
|
BASP1
|
brain abundant, membrane attached signal protein 1 |
| chr6_+_32811885 | 0.46 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr20_+_37353084 | 0.45 |
ENST00000217420.1
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr19_+_50706866 | 0.45 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr15_-_45480147 | 0.45 |
ENST00000560734.1
|
SHF
|
Src homology 2 domain containing F |
| chr13_+_95364963 | 0.45 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr12_-_131323754 | 0.44 |
ENST00000261653.6
|
STX2
|
syntaxin 2 |
| chr5_+_140254884 | 0.43 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr14_-_36983034 | 0.43 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr17_-_4463856 | 0.41 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr17_-_63556414 | 0.41 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr19_+_14544099 | 0.41 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr22_-_46373004 | 0.39 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr1_-_204654826 | 0.39 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr16_-_30107491 | 0.39 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr14_+_32546274 | 0.38 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr20_+_13976015 | 0.38 |
ENST00000217246.4
|
MACROD2
|
MACRO domain containing 2 |
| chr1_+_61547405 | 0.38 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr4_-_39640700 | 0.37 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr1_-_204654481 | 0.37 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr2_-_233792837 | 0.37 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr19_-_3062465 | 0.36 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr5_+_42423872 | 0.36 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr1_+_61548225 | 0.36 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr13_+_98795505 | 0.36 |
ENST00000319562.6
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr19_-_38806560 | 0.36 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr21_-_40685504 | 0.35 |
ENST00000380800.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr14_-_67982146 | 0.35 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr13_+_98795434 | 0.35 |
ENST00000376586.2
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr19_-_38806540 | 0.35 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_-_32801825 | 0.34 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr5_+_110559603 | 0.33 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr21_-_40685477 | 0.33 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr18_+_13218769 | 0.33 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr4_-_39640513 | 0.33 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr22_-_31741757 | 0.33 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_8954491 | 0.32 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr3_+_12838161 | 0.32 |
ENST00000456430.2
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr14_-_67981916 | 0.32 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr15_+_57668695 | 0.31 |
ENST00000281282.5
|
CGNL1
|
cingulin-like 1 |
| chr3_+_195943369 | 0.31 |
ENST00000296327.5
|
SLC51A
|
solute carrier family 51, alpha subunit |
| chr1_-_40367668 | 0.31 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr17_+_55334364 | 0.31 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr10_+_35416223 | 0.31 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr10_+_60272814 | 0.30 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr2_+_97481974 | 0.30 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr17_-_42276574 | 0.30 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr14_-_67981870 | 0.30 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr11_-_46142615 | 0.29 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr6_-_79787902 | 0.29 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr1_-_47407111 | 0.28 |
ENST00000371904.4
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr9_+_86595626 | 0.27 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr12_+_9067123 | 0.27 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr16_+_2802623 | 0.27 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr19_+_11457175 | 0.27 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr11_-_110583451 | 0.26 |
ENST00000260283.4
ENST00000528829.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr2_+_45878790 | 0.26 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr11_-_77531752 | 0.25 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr10_+_21823079 | 0.25 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_+_36273743 | 0.25 |
ENST00000373210.3
|
AGO4
|
argonaute RISC catalytic component 4 |
| chr5_+_139505520 | 0.25 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr3_+_111718173 | 0.25 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr7_-_73097741 | 0.25 |
ENST00000395176.2
|
DNAJC30
|
DnaJ (Hsp40) homolog, subfamily C, member 30 |
| chr3_-_113415441 | 0.25 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr19_-_49565254 | 0.24 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr11_+_17756279 | 0.24 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr3_-_27763803 | 0.24 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr9_+_125703282 | 0.24 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr2_+_232573208 | 0.24 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr3_+_184097905 | 0.24 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr11_-_102323740 | 0.24 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr5_+_102594403 | 0.23 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr6_-_119670919 | 0.23 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr7_+_30323923 | 0.23 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr20_-_61493115 | 0.23 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr7_-_100493482 | 0.23 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr11_-_72496976 | 0.23 |
ENST00000539138.1
ENST00000542989.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_+_88108381 | 0.23 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr5_-_111093081 | 0.23 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr14_+_32546145 | 0.23 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr22_+_39745930 | 0.22 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr12_-_131323719 | 0.22 |
ENST00000392373.2
|
STX2
|
syntaxin 2 |
| chr14_-_21493884 | 0.22 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr11_-_64612041 | 0.21 |
ENST00000342711.5
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr1_+_206730484 | 0.21 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr21_+_46825032 | 0.21 |
ENST00000400337.2
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr3_-_11610255 | 0.21 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr9_-_112260531 | 0.21 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr14_-_21493649 | 0.21 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr14_-_21493123 | 0.21 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr5_+_139175380 | 0.20 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr15_+_76352178 | 0.20 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr3_+_73045936 | 0.20 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr3_+_184098065 | 0.20 |
ENST00000348986.3
|
CHRD
|
chordin |
| chr8_+_30241934 | 0.20 |
ENST00000538486.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr17_+_65821780 | 0.20 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr3_+_38495333 | 0.20 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr10_-_52383644 | 0.20 |
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1 |
| chr10_-_15210615 | 0.20 |
ENST00000378150.1
|
NMT2
|
N-myristoyltransferase 2 |
| chr20_-_62710832 | 0.20 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr19_-_45661995 | 0.20 |
ENST00000438936.2
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr7_+_128312346 | 0.20 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr14_+_55738021 | 0.19 |
ENST00000313833.4
|
FBXO34
|
F-box protein 34 |
| chr11_-_46142948 | 0.19 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr6_+_31865552 | 0.19 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr14_-_89883412 | 0.19 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr14_+_90528109 | 0.19 |
ENST00000282146.4
|
KCNK13
|
potassium channel, subfamily K, member 13 |
| chr11_-_77531858 | 0.19 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr2_+_48757278 | 0.19 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr17_+_42634844 | 0.19 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr3_-_71774516 | 0.19 |
ENST00000425534.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr11_-_46142505 | 0.19 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr13_-_45151259 | 0.19 |
ENST00000493016.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr6_+_31683117 | 0.18 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr6_-_99395787 | 0.18 |
ENST00000369244.2
ENST00000229971.1 |
FBXL4
|
F-box and leucine-rich repeat protein 4 |
| chr12_+_26274917 | 0.18 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr16_+_50187556 | 0.18 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr12_+_68042495 | 0.18 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr9_-_84303269 | 0.18 |
ENST00000418319.1
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
| chr2_+_39893043 | 0.18 |
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A |
| chr2_+_139259324 | 0.18 |
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like |
| chr7_+_89841024 | 0.18 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr7_-_100493744 | 0.18 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr1_-_177133818 | 0.18 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr8_+_30241995 | 0.18 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr2_-_198650037 | 0.18 |
ENST00000392296.4
|
BOLL
|
boule-like RNA-binding protein |
| chr1_+_203765437 | 0.17 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr3_+_184097836 | 0.17 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr6_+_6588316 | 0.17 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr9_-_14313893 | 0.17 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr10_+_22610124 | 0.17 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr14_+_58765103 | 0.17 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_-_150675372 | 0.17 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr7_+_89841000 | 0.16 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr17_-_41985096 | 0.16 |
ENST00000269095.4
ENST00000523220.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr2_+_232573222 | 0.16 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr19_+_56154913 | 0.16 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr18_-_5895954 | 0.16 |
ENST00000581347.2
|
TMEM200C
|
transmembrane protein 200C |
| chr17_+_19912640 | 0.16 |
ENST00000395527.4
ENST00000583482.2 ENST00000583528.1 ENST00000583463.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr2_+_46926326 | 0.16 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr3_-_197282821 | 0.16 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_-_185826286 | 0.16 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chr19_+_8117636 | 0.15 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr1_+_206680879 | 0.15 |
ENST00000355294.4
ENST00000367117.3 |
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr17_-_79869340 | 0.15 |
ENST00000538936.2
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr17_-_42327236 | 0.15 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr6_+_24495067 | 0.15 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr5_+_139028510 | 0.15 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr14_-_36990354 | 0.15 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_-_11485922 | 0.15 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr5_+_61602055 | 0.14 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr2_-_160472952 | 0.14 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr5_+_139027877 | 0.14 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr5_-_73937244 | 0.14 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr17_-_7382834 | 0.14 |
ENST00000380599.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr11_-_73309228 | 0.14 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr15_+_85291785 | 0.14 |
ENST00000560079.2
|
ZNF592
|
zinc finger protein 592 |
| chr2_+_149402553 | 0.14 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr10_+_181418 | 0.14 |
ENST00000403354.1
ENST00000381607.4 ENST00000402736.1 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr14_-_51297837 | 0.14 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr11_-_82782861 | 0.14 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr16_+_69958887 | 0.14 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr10_-_121356007 | 0.14 |
ENST00000369093.2
ENST00000436547.2 |
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr1_+_87797351 | 0.14 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr1_-_153895377 | 0.14 |
ENST00000368655.4
|
GATAD2B
|
GATA zinc finger domain containing 2B |
| chr3_+_179370517 | 0.13 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chrX_-_19905703 | 0.13 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr11_+_118307179 | 0.13 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.2 | 0.6 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.2 | 0.6 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 | 0.8 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.4 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.4 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.6 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.5 | GO:2001074 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.3 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 2.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0033216 | ferric iron import(GO:0033216) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.9 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.3 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.6 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 2.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.6 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.2 | 0.8 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |