Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
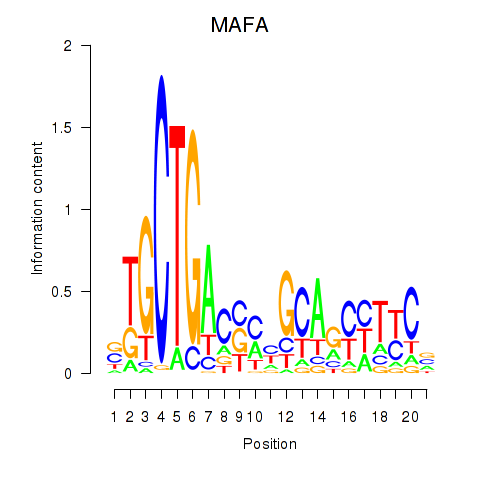
Results for MAFA
Z-value: 0.77
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAF bZIP transcription factor A |
Activity profile of MAFA motif
Sorted Z-values of MAFA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_39270725 | 2.11 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr22_-_28197486 | 1.96 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr11_+_118826999 | 1.72 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr2_+_33172221 | 1.71 |
ENST00000354476.3
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr10_+_120967072 | 1.68 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr2_+_85811525 | 1.56 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr11_+_72929319 | 1.52 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_72929402 | 1.47 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr20_+_306177 | 1.44 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr15_+_90744533 | 1.43 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr16_+_56598961 | 1.39 |
ENST00000219162.3
|
MT4
|
metallothionein 4 |
| chrX_-_106959631 | 1.33 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_-_51141196 | 1.26 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr6_+_7727030 | 1.14 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr11_-_66103932 | 1.11 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr19_-_43099070 | 1.10 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr17_-_39324424 | 1.08 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr19_-_4124079 | 1.03 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr11_-_66103867 | 1.02 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr10_-_79397391 | 1.01 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr20_+_306221 | 0.98 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr16_-_46865286 | 0.98 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr2_+_118846008 | 0.96 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr17_-_41623009 | 0.91 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr10_+_124221036 | 0.89 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chr10_-_29811456 | 0.88 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chrX_-_43741594 | 0.87 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr15_+_80696666 | 0.87 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr7_-_73133959 | 0.83 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr10_-_75676400 | 0.78 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr7_-_100860851 | 0.77 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_+_110754094 | 0.75 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr5_+_139739772 | 0.73 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr2_+_191745560 | 0.73 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr17_-_47925379 | 0.71 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr2_+_115199876 | 0.67 |
ENST00000436732.1
ENST00000410059.1 |
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr3_+_185304059 | 0.66 |
ENST00000427465.2
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr2_-_30143525 | 0.65 |
ENST00000431873.1
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr6_+_42896865 | 0.62 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr5_-_114505624 | 0.61 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr18_+_77623668 | 0.61 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr5_-_22853429 | 0.61 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr12_-_105478339 | 0.60 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr17_-_39254391 | 0.59 |
ENST00000333822.4
|
KRTAP4-8
|
keratin associated protein 4-8 |
| chr5_+_60933634 | 0.59 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr16_-_46865047 | 0.58 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr9_-_100000957 | 0.55 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr11_-_104817919 | 0.55 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chrX_+_68725084 | 0.55 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr9_-_28670283 | 0.54 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr8_+_85095769 | 0.54 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr19_+_18451391 | 0.54 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr8_+_85095497 | 0.53 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr11_+_107462471 | 0.52 |
ENST00000600612.1
|
AP000889.3
|
HCG2032453; Uncharacterized protein; cDNA FLJ25337 fis, clone TST00714 |
| chr1_-_38325256 | 0.52 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr2_+_191745535 | 0.50 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr12_-_21654479 | 0.49 |
ENST00000421138.2
ENST00000444129.2 ENST00000539672.1 ENST00000542432.1 ENST00000536964.1 ENST00000536240.1 ENST00000396093.3 ENST00000314748.6 |
RECQL
|
RecQ protein-like (DNA helicase Q1-like) |
| chr1_+_200842083 | 0.47 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chrX_-_63425561 | 0.46 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr17_-_76128488 | 0.46 |
ENST00000322914.3
|
TMC6
|
transmembrane channel-like 6 |
| chr19_-_2050852 | 0.45 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr10_-_79397479 | 0.44 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_+_21654714 | 0.43 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr10_+_135122906 | 0.42 |
ENST00000368554.4
|
ZNF511
|
zinc finger protein 511 |
| chr14_+_24600484 | 0.42 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr17_+_38465441 | 0.42 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr4_+_74606223 | 0.41 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr13_+_28194873 | 0.41 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr6_+_116692102 | 0.40 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr19_+_18451439 | 0.40 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr6_-_99797522 | 0.40 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr3_-_48754599 | 0.39 |
ENST00000413654.1
ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr3_+_42190714 | 0.39 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr21_+_41239243 | 0.38 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr9_+_2621798 | 0.37 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr9_+_33629119 | 0.37 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr22_+_23063100 | 0.37 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr9_+_2622085 | 0.36 |
ENST00000382099.2
|
VLDLR
|
very low density lipoprotein receptor |
| chr20_-_23066953 | 0.36 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr7_-_108168580 | 0.35 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr10_-_49701686 | 0.35 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr3_-_126277796 | 0.34 |
ENST00000318225.2
|
C3orf22
|
chromosome 3 open reading frame 22 |
| chr10_-_70231639 | 0.33 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr7_-_151433342 | 0.32 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_-_777467 | 0.32 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chr17_+_31318886 | 0.32 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr17_+_41363854 | 0.31 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr14_+_81421861 | 0.31 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr12_-_2027639 | 0.31 |
ENST00000586184.1
ENST00000587995.1 ENST00000585732.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr5_+_176853702 | 0.30 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr22_+_23161491 | 0.30 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr12_+_56546363 | 0.29 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr14_+_81421355 | 0.29 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr10_+_102505468 | 0.29 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chrX_-_92928557 | 0.29 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr22_+_22599075 | 0.29 |
ENST00000403807.3
|
VPREB1
|
pre-B lymphocyte 1 |
| chr17_-_10325261 | 0.28 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr4_-_10118348 | 0.27 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr8_-_63998590 | 0.26 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr16_-_79015739 | 0.25 |
ENST00000594986.1
|
PIH1
|
HCG1979943; Uncharacterized protein |
| chr17_+_7323634 | 0.25 |
ENST00000323675.3
|
SPEM1
|
spermatid maturation 1 |
| chr15_-_23086394 | 0.24 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr8_-_89339705 | 0.24 |
ENST00000286614.6
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted) |
| chr22_+_22697537 | 0.23 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr19_-_48547294 | 0.22 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr11_+_33563821 | 0.21 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr10_-_81708854 | 0.21 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr17_-_39728303 | 0.19 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chr15_-_90456114 | 0.18 |
ENST00000398333.3
|
C15orf38-AP3S2
|
C15orf38-AP3S2 readthrough |
| chr3_+_196439275 | 0.18 |
ENST00000296333.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr12_+_79258547 | 0.18 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr1_+_66458072 | 0.18 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr17_+_25936862 | 0.18 |
ENST00000582410.1
|
KSR1
|
kinase suppressor of ras 1 |
| chr2_+_103236004 | 0.17 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr11_+_107461948 | 0.17 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr11_-_70507901 | 0.17 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr12_+_79258444 | 0.17 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr16_+_56225248 | 0.15 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr20_-_31592113 | 0.15 |
ENST00000420875.1
ENST00000375519.2 ENST00000375523.3 ENST00000356173.3 |
SUN5
|
Sad1 and UNC84 domain containing 5 |
| chr15_-_90456156 | 0.13 |
ENST00000357484.5
|
C15orf38
|
chromosome 15 open reading frame 38 |
| chr11_-_57177586 | 0.13 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr7_-_135412925 | 0.13 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr22_+_22730353 | 0.12 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr20_-_30795511 | 0.12 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chrX_-_1511617 | 0.12 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr7_+_100860949 | 0.12 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr6_-_24489842 | 0.12 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr3_+_141121164 | 0.11 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr22_+_22599189 | 0.11 |
ENST00000302273.2
|
VPREB1
|
pre-B lymphocyte 1 |
| chr19_+_36103631 | 0.11 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr11_-_2182388 | 0.10 |
ENST00000421783.1
ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr19_+_33182823 | 0.10 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr6_+_22221010 | 0.10 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr1_-_32403370 | 0.10 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr4_+_2470664 | 0.09 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chr11_+_111411384 | 0.08 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr22_+_48972118 | 0.08 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr6_+_97372596 | 0.08 |
ENST00000369261.4
|
KLHL32
|
kelch-like family member 32 |
| chr13_-_103426081 | 0.07 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr19_-_50464338 | 0.07 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr4_-_151936865 | 0.06 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr13_-_103426112 | 0.05 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr17_+_18759612 | 0.05 |
ENST00000432893.2
ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr11_-_64660916 | 0.04 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr11_-_62783276 | 0.03 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr22_-_30662828 | 0.03 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr19_+_12949251 | 0.02 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr3_-_49823941 | 0.02 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr15_-_41047421 | 0.02 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chr17_-_3796334 | 0.02 |
ENST00000381771.2
ENST00000348335.2 ENST00000158166.5 |
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr3_+_196439170 | 0.01 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr3_+_107241882 | 0.01 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_74161977 | 0.01 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr15_-_23086259 | 0.01 |
ENST00000538684.1
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.3 | 0.8 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 0.3 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.2 | 1.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 2.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 2.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 3.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 1.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.2 | 0.8 | GO:0010807 | positive regulation of norepinephrine secretion(GO:0010701) regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.6 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.2 | 1.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 1.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.5 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.8 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 1.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.3 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 1.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 1.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 2.4 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 1.7 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 0.8 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 1.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 0.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 0.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 3.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 2.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.8 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |