Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
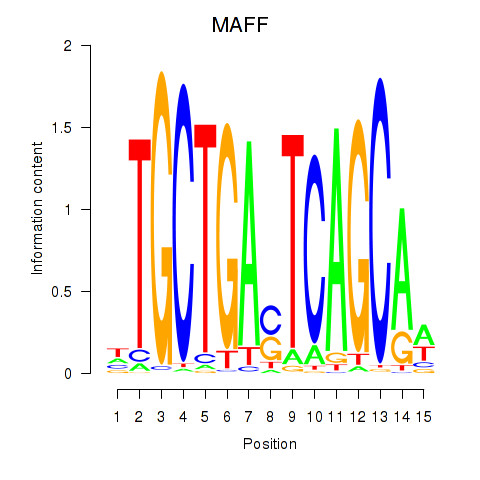
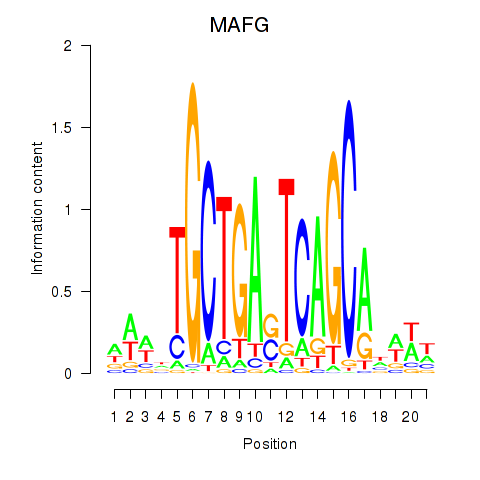
Results for MAFF_MAFG
Z-value: 1.35
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg19_v2_chr22_+_38609538_38609547 | -0.57 | 9.0e-04 | Click! |
| MAFG | hg19_v2_chr17_-_79885576_79885624 | -0.43 | 1.9e-02 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 21.53 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr1_+_85527987 | 9.96 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr19_+_5914213 | 9.36 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr12_+_111051902 | 8.47 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chrY_+_2709906 | 7.98 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr5_-_54468974 | 7.51 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr6_+_33043703 | 6.07 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr17_-_61512545 | 5.86 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr4_-_16077741 | 5.63 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr6_-_43478239 | 5.32 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr16_+_30211181 | 5.17 |
ENST00000395138.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr16_+_80574854 | 5.11 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr4_+_15480828 | 4.86 |
ENST00000389652.5
|
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr1_-_109655355 | 4.50 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr11_-_5248294 | 4.41 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr5_+_140019079 | 4.40 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr1_-_109655377 | 4.31 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr16_+_4838412 | 4.27 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr2_+_120189422 | 4.10 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr7_-_154863264 | 4.01 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr10_+_134150835 | 3.85 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr17_+_260097 | 3.65 |
ENST00000360127.6
ENST00000571106.1 ENST00000491373.1 |
C17orf97
|
chromosome 17 open reading frame 97 |
| chr9_-_138391692 | 3.65 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr12_-_71533055 | 3.64 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_17248418 | 3.49 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr15_+_90895471 | 3.43 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr2_-_220108309 | 3.40 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like |
| chr12_-_10282836 | 3.36 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_+_27076764 | 3.24 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_+_108308513 | 3.18 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr8_-_110620284 | 3.09 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr17_-_4689649 | 3.01 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chrX_-_55187531 | 2.92 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr17_-_4689727 | 2.84 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chrX_-_55187588 | 2.81 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr5_+_140602904 | 2.58 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr10_+_115803650 | 2.54 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr16_+_82068830 | 2.42 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr5_-_75013193 | 2.29 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr1_+_183774240 | 2.23 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr16_+_4838393 | 2.22 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr12_-_10282742 | 2.08 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr5_-_169626104 | 2.04 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr12_-_10282681 | 2.03 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr1_-_150669500 | 1.95 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr5_+_140019004 | 1.91 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr7_-_150777874 | 1.91 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr1_-_207095324 | 1.88 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr11_+_75428857 | 1.87 |
ENST00000198801.5
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_-_86849883 | 1.86 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr17_+_53343577 | 1.85 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr3_-_46506358 | 1.84 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr12_-_39837192 | 1.79 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr12_-_39836772 | 1.78 |
ENST00000541463.2
ENST00000361418.5 ENST00000544797.2 |
KIF21A
|
kinesin family member 21A |
| chr22_-_30867973 | 1.75 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr7_-_122526799 | 1.74 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr11_-_34938039 | 1.73 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr10_-_116444371 | 1.70 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr7_-_150777920 | 1.68 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr22_-_37172111 | 1.67 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr6_+_27215494 | 1.66 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr19_-_33555780 | 1.66 |
ENST00000254260.3
ENST00000400226.4 |
RHPN2
|
rhophilin, Rho GTPase binding protein 2 |
| chr7_-_150777949 | 1.64 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_+_44645700 | 1.64 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr4_+_74718906 | 1.63 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr11_-_119211525 | 1.61 |
ENST00000528368.1
|
C1QTNF5
|
C1q and tumor necrosis factor related protein 5 |
| chr22_-_37172191 | 1.60 |
ENST00000340630.5
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr16_+_616995 | 1.58 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chr21_+_42733870 | 1.57 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr4_-_101111615 | 1.56 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr7_-_99679324 | 1.55 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr6_+_30951487 | 1.54 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr11_-_34533257 | 1.54 |
ENST00000312319.2
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr1_-_157567868 | 1.52 |
ENST00000271532.1
|
FCRL4
|
Fc receptor-like 4 |
| chr1_+_145549203 | 1.49 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr3_-_46506563 | 1.48 |
ENST00000231751.4
|
LTF
|
lactotransferrin |
| chr5_-_13944652 | 1.48 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr16_-_25122785 | 1.46 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr5_+_139027877 | 1.45 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr18_+_61575200 | 1.40 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr12_+_51318513 | 1.39 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr16_-_28608364 | 1.36 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr16_+_66442411 | 1.36 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr11_-_114271139 | 1.35 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr3_-_57530051 | 1.34 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr5_-_111093167 | 1.33 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr18_-_52989525 | 1.32 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr19_+_44645731 | 1.30 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr5_-_42811986 | 1.28 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr18_-_53068911 | 1.27 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr11_+_124055923 | 1.26 |
ENST00000318666.6
|
OR10D3
|
olfactory receptor, family 10, subfamily D, member 3 (non-functional) |
| chrX_-_10544942 | 1.25 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr10_-_7513904 | 1.25 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr18_-_53069419 | 1.24 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr16_-_3350614 | 1.24 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr5_-_111092873 | 1.22 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr7_-_22234381 | 1.21 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr3_+_197687071 | 1.18 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr17_-_63822563 | 1.14 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr11_+_121447469 | 1.12 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr5_-_111093406 | 1.12 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr2_+_191273052 | 1.11 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr3_-_161090660 | 1.10 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr5_-_42812143 | 1.09 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr9_-_119449483 | 1.08 |
ENST00000288520.5
ENST00000358637.4 ENST00000341734.4 |
ASTN2
|
astrotactin 2 |
| chr6_-_159420780 | 1.07 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chrX_-_73512411 | 1.07 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr11_+_1855645 | 1.06 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr1_+_13521973 | 1.06 |
ENST00000327795.5
|
PRAMEF21
|
PRAME family member 21 |
| chr1_-_108231101 | 1.05 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr12_+_20968608 | 1.05 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_-_13544945 | 1.03 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr7_+_12610307 | 1.02 |
ENST00000297029.5
|
SCIN
|
scinderin |
| chr19_-_55791431 | 1.01 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_-_10491234 | 1.01 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr16_-_1429627 | 0.98 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr4_+_154125565 | 0.98 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr2_+_74648848 | 0.97 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr12_-_57522813 | 0.96 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr14_-_75530693 | 0.96 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr19_-_23578220 | 0.96 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr6_-_31681839 | 0.96 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr1_-_54872059 | 0.95 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr6_-_47277634 | 0.94 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr11_-_77791156 | 0.93 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr2_+_198570081 | 0.93 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr4_-_123843597 | 0.92 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr17_+_32646055 | 0.92 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chr6_+_159291090 | 0.92 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr1_-_27998689 | 0.92 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr1_+_22351977 | 0.91 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr6_+_88299833 | 0.90 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr6_-_152489484 | 0.89 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr10_-_100174900 | 0.89 |
ENST00000370575.4
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr5_+_130506475 | 0.89 |
ENST00000379380.4
|
LYRM7
|
LYR motif containing 7 |
| chr8_+_110099653 | 0.88 |
ENST00000311762.2
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr2_+_74781828 | 0.88 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr11_+_66276550 | 0.87 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr1_+_13742808 | 0.85 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr7_+_63774321 | 0.85 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr17_+_30771279 | 0.84 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr6_-_109330702 | 0.83 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr19_+_41816053 | 0.83 |
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97 |
| chr4_+_40337340 | 0.82 |
ENST00000310169.2
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
| chr22_-_20461786 | 0.82 |
ENST00000426804.1
|
RIMBP3
|
RIMS binding protein 3 |
| chr16_+_14802801 | 0.80 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chrX_+_30260054 | 0.80 |
ENST00000378982.2
|
MAGEB4
|
melanoma antigen family B, 4 |
| chr6_+_27215471 | 0.79 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr12_-_91573132 | 0.79 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr6_+_126112001 | 0.79 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_39421377 | 0.78 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr22_-_27014043 | 0.76 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr1_-_166845515 | 0.76 |
ENST00000367874.4
|
TADA1
|
transcriptional adaptor 1 |
| chr3_-_150264272 | 0.75 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr11_+_118978045 | 0.75 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr18_-_52989217 | 0.75 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr2_-_219858123 | 0.75 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chr9_+_71939488 | 0.74 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr9_-_130639997 | 0.74 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr14_+_55590646 | 0.73 |
ENST00000553493.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr2_-_174828892 | 0.71 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr18_-_53257027 | 0.70 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr4_-_1670632 | 0.70 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr7_+_89975979 | 0.69 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr6_-_43197189 | 0.69 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr13_+_108921977 | 0.68 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr13_-_53024725 | 0.68 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr2_+_128458514 | 0.67 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chrX_+_102883887 | 0.67 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr5_+_99871004 | 0.66 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr3_-_125802765 | 0.66 |
ENST00000514891.1
ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr21_+_17909594 | 0.66 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_133613937 | 0.66 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr22_-_19435209 | 0.65 |
ENST00000546308.1
ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA
C22orf39
|
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr7_+_100199800 | 0.65 |
ENST00000223061.5
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr7_-_5465045 | 0.65 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr19_+_37837185 | 0.65 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr7_+_139025875 | 0.64 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr5_+_89854595 | 0.64 |
ENST00000405460.2
|
GPR98
|
G protein-coupled receptor 98 |
| chr22_-_21905750 | 0.64 |
ENST00000433039.1
|
RIMBP3C
|
RIMS binding protein 3C |
| chr10_-_44144292 | 0.64 |
ENST00000374433.2
|
ZNF32
|
zinc finger protein 32 |
| chrX_+_129473916 | 0.64 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr17_-_79166176 | 0.63 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr4_-_140222358 | 0.62 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_-_28220002 | 0.62 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr2_+_163200848 | 0.62 |
ENST00000233612.4
|
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr5_-_111093081 | 0.62 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr5_-_111093340 | 0.62 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr18_-_40695604 | 0.62 |
ENST00000590910.1
ENST00000326695.5 ENST00000589109.1 ENST00000282028.4 |
RIT2
|
Ras-like without CAAX 2 |
| chr3_-_142720267 | 0.61 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr5_+_110409012 | 0.61 |
ENST00000379706.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr3_-_3221358 | 0.61 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr1_-_207095212 | 0.60 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr11_+_60048129 | 0.60 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr13_+_108922228 | 0.60 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr10_+_82300575 | 0.59 |
ENST00000313455.4
|
SH2D4B
|
SH2 domain containing 4B |
| chr2_+_163200598 | 0.59 |
ENST00000437150.2
ENST00000453113.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr8_-_95220775 | 0.58 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr11_+_60048004 | 0.58 |
ENST00000532114.1
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr7_+_75911902 | 0.58 |
ENST00000413003.1
|
SRRM3
|
serine/arginine repetitive matrix 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.4 | 5.6 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.1 | 3.3 | GO:0033212 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 1.1 | 4.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.9 | 3.5 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.8 | 2.5 | GO:0003099 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.7 | 5.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 1.1 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.4 | 7.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.3 | 1.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 0.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.3 | 1.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 0.9 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 3.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.3 | 0.8 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 1.5 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.3 | 1.3 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.2 | 1.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 2.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.2 | 28.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.7 | GO:0070269 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.2 | 0.9 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 8.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.5 | GO:0036446 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.7 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 2.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.6 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 1.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.5 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.7 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.6 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.9 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.6 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.5 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.4 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.1 | 1.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 1.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 1.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 6.1 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.1 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.7 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 0.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.2 | GO:0040010 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.5 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 1.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 7.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.3 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 13.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 5.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 2.8 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 3.5 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 2.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 5.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.4 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 1.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 1.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 4.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 4.8 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.0 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.3 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.7 | 13.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 6.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 3.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.5 | 3.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 29.2 | GO:0005844 | polysome(GO:0005844) |
| 0.3 | 3.1 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 0.9 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 6.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 5.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 0.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 0.8 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 11.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 1.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 0.5 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 3.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 3.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 2.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) clathrin complex(GO:0071439) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 1.5 | 5.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.9 | 7.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.9 | 5.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.8 | 2.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.8 | 3.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 1.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 10.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 5.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 2.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 29.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 3.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 1.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.7 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.3 | 1.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 0.8 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 6.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 6.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 2.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 0.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 1.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 6.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 0.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 3.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 4.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.9 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 2.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 4.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 3.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.5 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.1 | GO:0030276 | calcium-dependent phospholipid binding(GO:0005544) clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.6 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 1.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 7.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 29.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 6.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 6.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 5.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |