Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
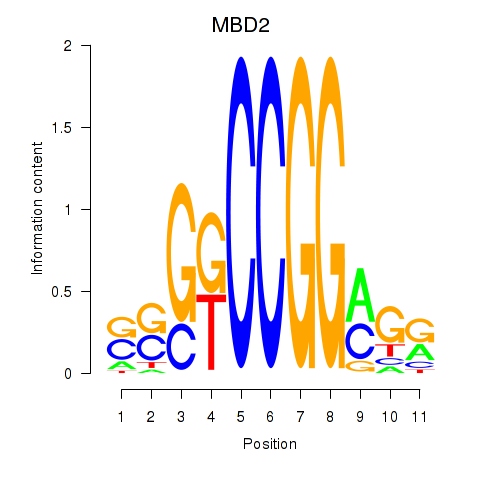
Results for MBD2
Z-value: 0.60
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51750948_51751059 | -0.15 | 4.2e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17659234 | 1.05 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr20_-_56284816 | 0.86 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr19_+_18496957 | 0.83 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr19_+_39904168 | 0.66 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_-_43596899 | 0.65 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr19_-_19051103 | 0.60 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr18_+_56530794 | 0.54 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr12_-_117318788 | 0.49 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr17_+_17942684 | 0.45 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr19_+_35491174 | 0.44 |
ENST00000317991.5
ENST00000504615.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr19_+_56152262 | 0.43 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr14_+_74417192 | 0.42 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr2_-_129076151 | 0.41 |
ENST00000259241.6
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr7_-_94285402 | 0.39 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr20_-_590944 | 0.39 |
ENST00000246080.3
|
TCF15
|
transcription factor 15 (basic helix-loop-helix) |
| chr7_-_94285472 | 0.39 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr1_-_43232649 | 0.39 |
ENST00000372526.2
ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr3_-_197024394 | 0.38 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_+_122064673 | 0.38 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr7_-_94285511 | 0.37 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr14_-_77787198 | 0.37 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr5_-_72744336 | 0.36 |
ENST00000499003.3
|
FOXD1
|
forkhead box D1 |
| chr19_+_35491330 | 0.36 |
ENST00000411896.2
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr6_+_4776580 | 0.36 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chr3_-_196295437 | 0.35 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr3_-_48471454 | 0.34 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr10_-_17659357 | 0.33 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr5_+_14143728 | 0.33 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr15_+_40763150 | 0.33 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr3_-_27525865 | 0.33 |
ENST00000445684.1
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr17_-_27278445 | 0.32 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr3_-_27525826 | 0.32 |
ENST00000454389.1
ENST00000440156.1 ENST00000437179.1 ENST00000446700.1 ENST00000455077.1 ENST00000435667.2 ENST00000388777.4 ENST00000425128.2 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr3_-_128206759 | 0.31 |
ENST00000430265.2
|
GATA2
|
GATA binding protein 2 |
| chr8_+_30241934 | 0.31 |
ENST00000538486.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr9_+_34989638 | 0.30 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_-_25012115 | 0.30 |
ENST00000446003.1
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chrX_+_68725084 | 0.30 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr20_-_48532046 | 0.30 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chrX_-_152939252 | 0.30 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr20_-_46415341 | 0.29 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr14_+_53019993 | 0.29 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr17_+_65374075 | 0.29 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_-_165325939 | 0.29 |
ENST00000342310.3
|
LMX1A
|
LIM homeobox transcription factor 1, alpha |
| chr21_-_44495919 | 0.28 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr4_-_16228083 | 0.28 |
ENST00000399920.3
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr6_+_125474992 | 0.28 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr15_+_80215477 | 0.28 |
ENST00000542003.1
|
C15ORF37
|
C15orf37 protein; Uncharacterized protein |
| chr14_+_53019822 | 0.28 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr2_-_54014127 | 0.27 |
ENST00000394717.2
|
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr2_-_208031542 | 0.27 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_196295468 | 0.27 |
ENST00000332629.5
ENST00000433160.1 |
WDR53
|
WD repeat domain 53 |
| chr11_-_64684672 | 0.27 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr20_-_46415297 | 0.27 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr12_-_120554534 | 0.27 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr17_-_7141490 | 0.26 |
ENST00000574236.1
ENST00000572789.1 |
PHF23
|
PHD finger protein 23 |
| chr19_-_41222775 | 0.26 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr3_+_133292759 | 0.26 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr15_+_101142722 | 0.26 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr3_+_133292851 | 0.26 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr18_-_31803169 | 0.26 |
ENST00000590712.1
|
NOL4
|
nucleolar protein 4 |
| chr3_-_98620500 | 0.26 |
ENST00000326840.6
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr5_-_180076580 | 0.26 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr9_+_35829208 | 0.25 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr7_-_112579869 | 0.25 |
ENST00000297145.4
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr14_+_21538517 | 0.25 |
ENST00000298693.3
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr5_+_133861790 | 0.25 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr7_+_73082152 | 0.25 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr6_-_91296737 | 0.24 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr6_-_91296602 | 0.24 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr2_+_173292390 | 0.24 |
ENST00000442250.1
ENST00000458358.1 ENST00000409080.1 |
ITGA6
|
integrin, alpha 6 |
| chr9_+_124413873 | 0.24 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr17_+_17942594 | 0.24 |
ENST00000268719.4
|
GID4
|
GID complex subunit 4 |
| chr7_-_19157248 | 0.23 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr11_+_67056755 | 0.23 |
ENST00000511455.2
ENST00000308440.6 |
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr1_-_6545502 | 0.23 |
ENST00000535355.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr3_+_33155525 | 0.23 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr2_-_54014055 | 0.23 |
ENST00000263634.3
ENST00000406687.1 |
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr3_+_88108381 | 0.23 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr11_+_67056867 | 0.23 |
ENST00000514166.1
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr14_+_77787227 | 0.22 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr6_+_125475335 | 0.22 |
ENST00000532429.1
ENST00000534199.1 |
TPD52L1
|
tumor protein D52-like 1 |
| chr5_+_122847908 | 0.22 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr11_+_67033881 | 0.22 |
ENST00000308595.5
ENST00000526285.1 |
ADRBK1
|
adrenergic, beta, receptor kinase 1 |
| chr7_+_94285637 | 0.21 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr3_-_88108212 | 0.21 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr18_-_34408802 | 0.21 |
ENST00000590842.1
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr2_+_48541776 | 0.21 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr2_+_11052054 | 0.21 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chrX_+_24167746 | 0.21 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr14_+_21538429 | 0.20 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr16_-_23160591 | 0.20 |
ENST00000219689.7
|
USP31
|
ubiquitin specific peptidase 31 |
| chr12_-_120554622 | 0.20 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr17_-_42441204 | 0.20 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr11_-_68609377 | 0.20 |
ENST00000265641.5
ENST00000376618.2 |
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr11_-_89224488 | 0.20 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr3_+_133292574 | 0.19 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr16_-_2059797 | 0.19 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr2_-_219537134 | 0.19 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chr9_+_129376722 | 0.19 |
ENST00000526117.1
ENST00000373474.4 ENST00000355497.5 ENST00000425646.2 ENST00000561065.1 |
LMX1B
|
LIM homeobox transcription factor 1, beta |
| chr4_+_2061119 | 0.19 |
ENST00000423729.2
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative) |
| chr3_-_48470838 | 0.19 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chrX_+_24167828 | 0.19 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr6_-_45983549 | 0.19 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr5_+_153570285 | 0.19 |
ENST00000425427.2
ENST00000297107.6 |
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr2_+_46769798 | 0.19 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr2_+_208576259 | 0.18 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr10_+_75545391 | 0.18 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr11_+_65408273 | 0.18 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr3_-_149688896 | 0.18 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr10_-_92617437 | 0.18 |
ENST00000336152.3
ENST00000277874.6 ENST00000371719.2 |
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
| chr2_+_9346892 | 0.18 |
ENST00000281419.3
ENST00000315273.4 |
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr5_-_171433579 | 0.18 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr22_-_19419205 | 0.18 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr17_+_6544356 | 0.18 |
ENST00000574838.1
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr1_-_247495045 | 0.18 |
ENST00000294753.4
ENST00000366498.2 |
ZNF496
|
zinc finger protein 496 |
| chr7_-_17980091 | 0.18 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr3_-_124774802 | 0.18 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr10_+_75545329 | 0.18 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr8_+_30241995 | 0.18 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr14_+_102829300 | 0.17 |
ENST00000359520.7
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr17_+_6544328 | 0.17 |
ENST00000570330.1
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr8_-_67525524 | 0.17 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr11_-_2160611 | 0.17 |
ENST00000416167.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_112930387 | 0.17 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chrX_-_134049233 | 0.17 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr18_-_45457192 | 0.17 |
ENST00000586514.1
ENST00000591214.1 ENST00000589877.1 |
SMAD2
|
SMAD family member 2 |
| chr5_-_171433819 | 0.17 |
ENST00000296933.6
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr2_+_173292301 | 0.17 |
ENST00000264106.6
ENST00000375221.2 ENST00000343713.4 |
ITGA6
|
integrin, alpha 6 |
| chrX_-_152939133 | 0.16 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr9_+_35605274 | 0.16 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr1_-_31712401 | 0.16 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr10_+_126490354 | 0.16 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr6_-_99797522 | 0.16 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr16_+_3019552 | 0.16 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr12_-_48213568 | 0.16 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr16_-_2059748 | 0.16 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr11_+_35965531 | 0.16 |
ENST00000528989.1
ENST00000524419.1 ENST00000315571.5 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr20_-_48532019 | 0.16 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr19_+_6739662 | 0.16 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr10_+_105253661 | 0.15 |
ENST00000369780.4
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr2_-_220408430 | 0.15 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr10_-_92617671 | 0.15 |
ENST00000371721.3
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
| chr19_-_17356697 | 0.15 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr22_+_25348671 | 0.15 |
ENST00000406486.4
|
KIAA1671
|
KIAA1671 |
| chr12_-_50222187 | 0.15 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr1_-_160232312 | 0.15 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr19_-_10341948 | 0.15 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr17_+_48638371 | 0.15 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_+_72766651 | 0.15 |
ENST00000379887.4
|
ARIH1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr11_+_67055986 | 0.15 |
ENST00000447274.2
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr19_-_11308190 | 0.15 |
ENST00000586659.1
ENST00000592903.1 ENST00000589359.1 ENST00000588724.1 ENST00000432929.2 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr16_+_3019246 | 0.15 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr1_-_156252590 | 0.15 |
ENST00000361813.5
ENST00000368267.5 |
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr3_+_120461484 | 0.15 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr19_-_11373128 | 0.15 |
ENST00000294618.7
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr11_+_65383227 | 0.14 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr20_-_30311703 | 0.14 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr12_+_27396901 | 0.14 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr20_-_30310797 | 0.14 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr14_-_99737822 | 0.14 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_-_154531095 | 0.14 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr17_+_48133459 | 0.14 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr19_-_4124079 | 0.14 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr11_+_66025938 | 0.13 |
ENST00000394066.2
|
KLC2
|
kinesin light chain 2 |
| chr20_-_50808236 | 0.13 |
ENST00000361387.2
|
ZFP64
|
ZFP64 zinc finger protein |
| chr17_+_65373531 | 0.13 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_+_3689325 | 0.13 |
ENST00000444870.2
ENST00000452264.1 |
SMIM1
|
small integral membrane protein 1 (Vel blood group) |
| chr1_+_4714792 | 0.13 |
ENST00000378190.3
|
AJAP1
|
adherens junctions associated protein 1 |
| chr12_+_107168418 | 0.13 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr19_-_17137625 | 0.13 |
ENST00000443236.1
ENST00000388925.4 |
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr5_+_153570319 | 0.13 |
ENST00000377661.2
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr5_+_176237478 | 0.13 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr19_+_50353944 | 0.13 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chrX_+_9983602 | 0.13 |
ENST00000380861.4
|
WWC3
|
WWC family member 3 |
| chr11_+_111473108 | 0.12 |
ENST00000304987.3
|
SIK2
|
salt-inducible kinase 2 |
| chr20_-_61885826 | 0.12 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr7_-_103848405 | 0.12 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr12_-_124457257 | 0.12 |
ENST00000545891.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr11_-_89224508 | 0.12 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr17_-_27949911 | 0.12 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr19_+_56154913 | 0.12 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr2_-_9695847 | 0.12 |
ENST00000310823.3
ENST00000497134.1 |
ADAM17
|
ADAM metallopeptidase domain 17 |
| chr3_+_49591881 | 0.12 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr17_+_46126135 | 0.12 |
ENST00000361665.3
ENST00000585062.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr10_+_128593978 | 0.12 |
ENST00000280333.6
|
DOCK1
|
dedicator of cytokinesis 1 |
| chr14_-_102829051 | 0.12 |
ENST00000536961.2
ENST00000541568.2 ENST00000216756.6 |
CINP
|
cyclin-dependent kinase 2 interacting protein |
| chr19_-_2151523 | 0.11 |
ENST00000350812.6
ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr1_-_200379129 | 0.11 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr2_+_219824357 | 0.11 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr6_+_1610681 | 0.11 |
ENST00000380874.2
|
FOXC1
|
forkhead box C1 |
| chr12_+_29542557 | 0.11 |
ENST00000550906.1
|
OVCH1-AS1
|
OVCH1 antisense RNA 1 |
| chr17_+_80477571 | 0.11 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr11_+_43380459 | 0.11 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr5_-_156569850 | 0.11 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr2_+_220299547 | 0.11 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_+_171785824 | 0.11 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr11_-_73471655 | 0.11 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr6_+_84569359 | 0.11 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr12_+_107168342 | 0.11 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr15_+_83654950 | 0.10 |
ENST00000304191.3
|
FAM103A1
|
family with sequence similarity 103, member A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.3 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.4 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.8 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0022009 | chronological cell aging(GO:0001300) establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |