Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
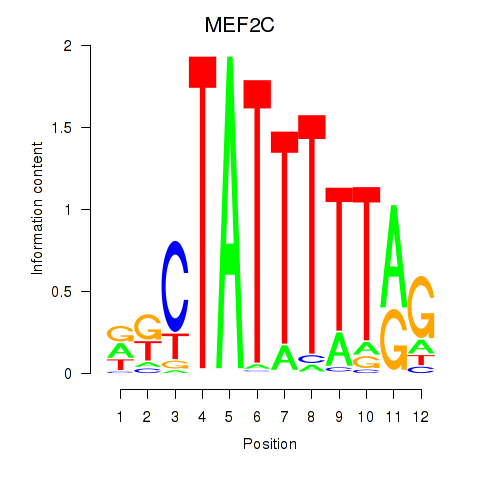
Results for MEF2C
Z-value: 0.71
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88119580_88119605 | -0.29 | 1.2e-01 | Click! |
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_8543533 | 3.72 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_+_8543393 | 2.66 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_+_8543561 | 2.56 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr15_-_70994612 | 1.97 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_+_143013198 | 1.44 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_+_123038689 | 1.22 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr4_-_143227088 | 1.21 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr14_-_94421923 | 1.15 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr7_-_113559104 | 1.10 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr12_-_111358372 | 0.99 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chrX_+_135279179 | 0.99 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135278908 | 0.81 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr14_+_76776957 | 0.77 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr1_+_74701062 | 0.74 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr3_+_50316458 | 0.74 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr3_+_99357319 | 0.73 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr3_+_35683651 | 0.72 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_235405679 | 0.69 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr2_-_208031542 | 0.69 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_159570722 | 0.66 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr11_+_69924639 | 0.63 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr1_+_90098606 | 0.63 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr6_-_76203454 | 0.59 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr19_-_49576198 | 0.59 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr14_+_24540731 | 0.58 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr9_+_116327326 | 0.57 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_-_186697044 | 0.57 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_1942580 | 0.56 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chrX_-_50386648 | 0.55 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr10_-_7513904 | 0.55 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr2_-_208030647 | 0.54 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_+_71249071 | 0.53 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr3_+_154797877 | 0.52 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr1_-_173174681 | 0.52 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr2_-_190044480 | 0.49 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_-_201391149 | 0.49 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr4_+_37892682 | 0.49 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr6_-_76203345 | 0.48 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr8_+_1993152 | 0.45 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr15_+_67418047 | 0.45 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr8_+_1993173 | 0.43 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr16_+_12059050 | 0.42 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_143767428 | 0.42 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr10_-_3827417 | 0.42 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chr20_-_44519839 | 0.41 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr15_+_59730348 | 0.41 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr7_+_129906660 | 0.40 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr13_-_30881134 | 0.40 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr16_+_12058961 | 0.39 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_57547454 | 0.39 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr1_-_12677714 | 0.39 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr19_-_51220176 | 0.38 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr7_-_121944491 | 0.38 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr16_+_30075463 | 0.38 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr5_-_95018660 | 0.37 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr4_-_57547870 | 0.36 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr4_+_169418255 | 0.36 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_26348259 | 0.36 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr3_+_138067666 | 0.36 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chrX_+_18725758 | 0.36 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr17_-_9694614 | 0.35 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr2_+_168043793 | 0.35 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr5_-_131347501 | 0.34 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_+_154002189 | 0.34 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr3_+_4535155 | 0.34 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr17_-_10276319 | 0.34 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr7_+_120629653 | 0.33 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_30075783 | 0.33 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr8_-_7309887 | 0.33 |
ENST00000458665.1
ENST00000528168.1 |
SPAG11B
|
sperm associated antigen 11B |
| chr8_-_49833978 | 0.33 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr16_+_30075595 | 0.33 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_+_4535025 | 0.32 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr22_-_36031181 | 0.32 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr10_+_71562180 | 0.32 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr11_+_1940925 | 0.31 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr3_-_112360116 | 0.31 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr3_+_138067521 | 0.31 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr16_-_31439735 | 0.30 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr6_-_123957942 | 0.30 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr5_-_11588907 | 0.30 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_-_108248169 | 0.30 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr11_+_1940786 | 0.30 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr7_-_143956815 | 0.30 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr15_+_68582544 | 0.29 |
ENST00000566008.1
|
FEM1B
|
fem-1 homolog b (C. elegans) |
| chr11_+_19799327 | 0.29 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr2_-_231825668 | 0.29 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr17_-_34625719 | 0.28 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr3_+_138067314 | 0.28 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr6_+_34204642 | 0.28 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr3_-_149510553 | 0.27 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr8_+_107282368 | 0.27 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr2_+_145780739 | 0.26 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_-_46904918 | 0.26 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr11_+_57308979 | 0.26 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr17_-_34524157 | 0.26 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr8_-_49834299 | 0.26 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr15_-_83474806 | 0.26 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr3_-_46904946 | 0.26 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr7_-_44105158 | 0.26 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr2_+_182850743 | 0.25 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr21_+_35552978 | 0.25 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr14_-_57277178 | 0.25 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr1_+_156095951 | 0.25 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr4_+_37962018 | 0.25 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr12_-_102874102 | 0.25 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr7_+_142457315 | 0.25 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr14_-_57277163 | 0.25 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr10_+_71561630 | 0.25 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_-_123958141 | 0.25 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr3_-_4927447 | 0.24 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr10_+_71561704 | 0.24 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_-_1389228 | 0.24 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr12_-_57644952 | 0.24 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr11_+_128563652 | 0.24 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr21_+_17909594 | 0.24 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_100643765 | 0.24 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr7_+_18535346 | 0.23 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr17_-_1389419 | 0.23 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr19_+_6739662 | 0.23 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr4_-_143226979 | 0.22 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr6_-_161085291 | 0.22 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr10_+_71561649 | 0.22 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr7_-_82073031 | 0.22 |
ENST00000356253.5
ENST00000423588.1 |
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr14_+_39735411 | 0.21 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr9_-_134585221 | 0.21 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chrX_-_30327495 | 0.21 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr4_-_186696425 | 0.21 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_11589131 | 0.21 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr2_+_148778570 | 0.21 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr5_-_66492562 | 0.21 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr3_-_137834436 | 0.20 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr8_-_72268889 | 0.20 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_+_55095264 | 0.20 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr16_+_12059091 | 0.20 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_+_143632259 | 0.19 |
ENST00000408955.2
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2 |
| chrX_-_15683147 | 0.19 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr1_+_230193521 | 0.19 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr7_-_82073109 | 0.19 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr8_+_145162629 | 0.19 |
ENST00000323662.8
|
KIAA1875
|
KIAA1875 |
| chr8_-_42234745 | 0.19 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr12_-_40013553 | 0.19 |
ENST00000308666.3
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr16_+_84328252 | 0.19 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr9_-_215744 | 0.18 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr22_-_37215523 | 0.18 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr1_+_198608146 | 0.18 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_24438664 | 0.18 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr8_+_7716700 | 0.18 |
ENST00000454911.2
ENST00000326625.5 |
SPAG11A
|
sperm associated antigen 11A |
| chr15_+_40531243 | 0.18 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr9_+_5231413 | 0.18 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr17_-_79881408 | 0.18 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr20_-_54580523 | 0.18 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chr11_-_85780853 | 0.18 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr17_-_72855989 | 0.17 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr19_+_35630344 | 0.17 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chrX_+_37545012 | 0.17 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr6_+_44194762 | 0.17 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr11_+_1860200 | 0.17 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr20_+_44035847 | 0.16 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_+_7382745 | 0.16 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chrX_+_131157609 | 0.16 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr3_+_141106643 | 0.16 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_+_161123270 | 0.16 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chrX_+_105192423 | 0.16 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr10_+_120116527 | 0.16 |
ENST00000445161.1
|
LINC00867
|
long intergenic non-protein coding RNA 867 |
| chr11_-_47400062 | 0.16 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr10_-_96829246 | 0.15 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr2_+_30569506 | 0.15 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr1_-_115238207 | 0.15 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr7_+_18535893 | 0.15 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr8_-_33457453 | 0.15 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr7_-_124569991 | 0.15 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr3_+_42727011 | 0.15 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr1_+_145413268 | 0.15 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr4_+_100495864 | 0.15 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr3_+_36421826 | 0.15 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr3_-_147124547 | 0.15 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr15_+_40531621 | 0.15 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr16_+_84328429 | 0.14 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr4_+_169418195 | 0.14 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_+_118083919 | 0.14 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr2_-_151344172 | 0.14 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr22_+_32340447 | 0.14 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr10_-_29923893 | 0.14 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr7_+_95115210 | 0.13 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr3_+_135741576 | 0.13 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr12_-_131200810 | 0.13 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr17_+_4855053 | 0.13 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr8_-_95565673 | 0.13 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr10_-_3827371 | 0.13 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr3_+_54157480 | 0.13 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr12_-_122240792 | 0.13 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chrX_-_109683446 | 0.13 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr22_+_40342819 | 0.12 |
ENST00000407075.3
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr8_-_72268968 | 0.12 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_-_71532339 | 0.12 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr18_-_35145981 | 0.12 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr1_-_40042416 | 0.12 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr21_+_39628852 | 0.12 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_67584174 | 0.12 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr18_-_35145728 | 0.12 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 0.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 0.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 1.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.5 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.3 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.2 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.4 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0042668 | inhibition of neuroepithelial cell differentiation(GO:0002085) trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 2.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 1.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 1.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0051096 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.0 | 0.3 | GO:0010811 | regulation of cell-substrate adhesion(GO:0010810) positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.0 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.7 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 1.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.5 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.4 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 1.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.4 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 9.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |