Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for MEF2D_MEF2A
Z-value: 0.56
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
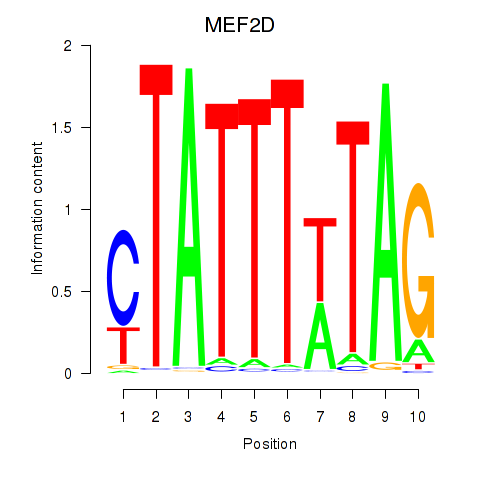
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
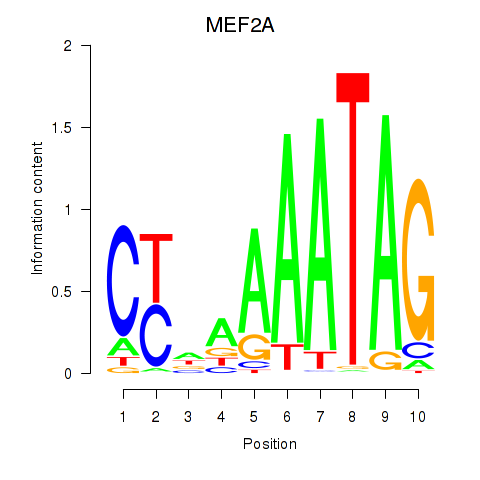
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg19_v2_chr1_-_156460391_156460417 | 0.15 | 4.3e-01 | Click! |
| MEF2A | hg19_v2_chr15_+_100106244_100106292 | -0.11 | 5.6e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_131200810 | 1.12 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr2_+_233497931 | 0.90 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr7_-_113559104 | 0.73 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr3_+_35683651 | 0.66 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_121944491 | 0.60 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr14_+_24540731 | 0.55 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr11_-_19223523 | 0.53 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr3_+_138067666 | 0.52 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr17_+_37821593 | 0.52 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr17_+_34538310 | 0.51 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr1_-_201391149 | 0.50 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr3_-_108248169 | 0.49 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr9_-_104145795 | 0.49 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr1_-_25756638 | 0.48 |
ENST00000349320.3
|
RHCE
|
Rh blood group, CcEe antigens |
| chr3_+_138067314 | 0.47 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr17_-_72855989 | 0.47 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr3_+_138067521 | 0.46 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr17_+_34639793 | 0.45 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr2_-_211168332 | 0.44 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr17_+_34640031 | 0.44 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr7_+_143013198 | 0.43 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr8_+_1993152 | 0.42 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr4_-_23891658 | 0.42 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr17_+_7184986 | 0.42 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr8_+_1993173 | 0.42 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr12_-_45269430 | 0.40 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr14_+_76776957 | 0.39 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr10_-_75415825 | 0.39 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr11_+_1942580 | 0.35 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr10_+_118187379 | 0.35 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chrX_-_21776281 | 0.35 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr8_+_7716700 | 0.34 |
ENST00000454911.2
ENST00000326625.5 |
SPAG11A
|
sperm associated antigen 11A |
| chr6_-_33160231 | 0.33 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr8_+_95565947 | 0.33 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr16_-_31439735 | 0.33 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr14_-_65409502 | 0.33 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr5_+_155753745 | 0.32 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr6_-_55739542 | 0.32 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr1_+_95583479 | 0.31 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr3_+_99357319 | 0.31 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr11_+_129245796 | 0.31 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr2_+_168043793 | 0.31 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr2_-_190044480 | 0.31 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr2_-_165424973 | 0.30 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr7_+_18535893 | 0.30 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr9_+_116327326 | 0.30 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_+_120056939 | 0.30 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr2_-_211179883 | 0.30 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr1_-_152131703 | 0.30 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr13_-_46756351 | 0.29 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr5_+_53751445 | 0.29 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr10_-_75410771 | 0.29 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr11_+_128563652 | 0.28 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_88367299 | 0.26 |
ENST00000419482.2
ENST00000444564.2 |
SMYD1
|
SET and MYND domain containing 1 |
| chr6_+_44194762 | 0.26 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr2_+_220299547 | 0.26 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chrX_-_15332665 | 0.25 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_+_150254936 | 0.25 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr17_-_34345002 | 0.24 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr8_-_7309887 | 0.24 |
ENST00000458665.1
ENST00000528168.1 |
SPAG11B
|
sperm associated antigen 11B |
| chr17_-_34344991 | 0.24 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr7_-_37488834 | 0.24 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_-_65409438 | 0.24 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_-_31637560 | 0.23 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr15_-_70994612 | 0.23 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_-_219696519 | 0.23 |
ENST00000545803.1
ENST00000430489.1 ENST00000392098.3 |
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr2_+_33661382 | 0.23 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr6_-_76203345 | 0.23 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_-_100643765 | 0.23 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr18_+_12407895 | 0.22 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr5_-_16509101 | 0.22 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_+_42248063 | 0.22 |
ENST00000293414.1
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr17_-_27949911 | 0.21 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr3_+_8543533 | 0.21 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr7_+_99775366 | 0.21 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr1_-_169703203 | 0.21 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr19_-_18209626 | 0.21 |
ENST00000598019.2
ENST00000594176.1 ENST00000600835.2 |
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr1_+_114522049 | 0.20 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr15_-_83474806 | 0.20 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr4_+_37892682 | 0.20 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr21_-_34186006 | 0.19 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr1_+_26348259 | 0.19 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr7_+_99775520 | 0.19 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr12_-_371994 | 0.19 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr9_+_129677123 | 0.19 |
ENST00000373436.1
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr6_+_123038689 | 0.19 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr3_+_50316458 | 0.19 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr16_+_84328252 | 0.19 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr6_-_76203454 | 0.19 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr16_+_84328429 | 0.19 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chrX_+_9217932 | 0.18 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chr17_-_1389228 | 0.18 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr2_-_219696511 | 0.18 |
ENST00000439262.2
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr12_+_15699286 | 0.18 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr20_-_44519839 | 0.17 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr1_+_203097407 | 0.17 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr1_-_116383322 | 0.17 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr17_-_10421853 | 0.17 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr1_+_145413268 | 0.17 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr10_-_3827371 | 0.17 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr1_+_182584314 | 0.17 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr8_-_42234745 | 0.17 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr18_+_13611431 | 0.16 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_1940925 | 0.16 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr19_-_47975417 | 0.16 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr7_+_30951461 | 0.16 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr4_-_122302163 | 0.16 |
ENST00000394427.2
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chrX_+_135570046 | 0.16 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr1_+_175036966 | 0.16 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr3_+_36421826 | 0.15 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr6_+_159291090 | 0.15 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr6_+_132129151 | 0.15 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_+_35258592 | 0.15 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr8_+_74903580 | 0.15 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr11_+_124481361 | 0.15 |
ENST00000284288.2
|
PANX3
|
pannexin 3 |
| chr6_+_159290917 | 0.14 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr16_+_12059091 | 0.14 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr11_-_47400078 | 0.14 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_+_39125365 | 0.14 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr14_+_39735411 | 0.14 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr15_-_99057551 | 0.14 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr6_-_45983581 | 0.14 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr10_-_45474237 | 0.14 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr4_-_143226979 | 0.13 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_154002189 | 0.13 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr6_-_27806117 | 0.13 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr8_+_27183033 | 0.13 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr5_-_150521192 | 0.13 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chrX_-_63450480 | 0.13 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr1_-_201390846 | 0.13 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr8_+_27182862 | 0.13 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr11_-_62457371 | 0.13 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr14_+_56046914 | 0.13 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr12_+_81101277 | 0.13 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr10_+_71562180 | 0.13 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr9_+_100263912 | 0.13 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr13_-_103346854 | 0.13 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr7_-_105332084 | 0.13 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_+_35721182 | 0.13 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr20_+_44657845 | 0.13 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr11_+_57308979 | 0.12 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr4_+_119809984 | 0.12 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr4_+_88529681 | 0.12 |
ENST00000399271.1
|
DSPP
|
dentin sialophosphoprotein |
| chr12_+_80750134 | 0.12 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chrX_-_50386648 | 0.12 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr17_-_10325261 | 0.12 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr18_-_35145728 | 0.12 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr17_-_1389419 | 0.12 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr14_-_57277178 | 0.12 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr18_-_35145981 | 0.12 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chrX_+_105192423 | 0.12 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr2_+_220144052 | 0.12 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_150245177 | 0.12 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr17_-_38821373 | 0.12 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr19_+_3933085 | 0.12 |
ENST00000168977.2
ENST00000599576.1 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr6_+_136172820 | 0.12 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr4_+_8321882 | 0.12 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr12_-_45270151 | 0.12 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr21_+_17909594 | 0.12 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_111358372 | 0.11 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr5_-_131347501 | 0.11 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr11_-_47399942 | 0.11 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr6_-_116833500 | 0.11 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr1_+_151129103 | 0.11 |
ENST00000368910.3
|
TNFAIP8L2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr18_-_44181442 | 0.11 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr6_-_169654139 | 0.11 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr12_-_99548270 | 0.11 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_8543393 | 0.11 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr4_+_119810134 | 0.11 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr8_-_27115931 | 0.11 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr2_+_145780739 | 0.11 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr2_-_29297127 | 0.11 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr11_+_112832202 | 0.11 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr8_-_27850141 | 0.11 |
ENST00000524352.1
|
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr7_-_44105158 | 0.10 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr6_+_152128686 | 0.10 |
ENST00000206249.3
|
ESR1
|
estrogen receptor 1 |
| chr12_-_45270077 | 0.10 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr5_+_67584174 | 0.10 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_-_131347306 | 0.10 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_+_123488124 | 0.10 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr10_+_88428370 | 0.10 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr17_-_10276319 | 0.10 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr3_-_57326704 | 0.10 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr3_+_132843652 | 0.10 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr14_+_100150622 | 0.10 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr2_+_220143989 | 0.10 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr17_-_41132410 | 0.09 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_+_68165657 | 0.09 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_204801471 | 0.09 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr13_+_76210448 | 0.09 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr18_+_13611763 | 0.09 |
ENST00000585931.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr1_-_99470558 | 0.09 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr8_-_70745575 | 0.09 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr10_+_32856764 | 0.09 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr1_-_23751189 | 0.09 |
ENST00000374601.3
ENST00000450454.2 |
TCEA3
|
transcription elongation factor A (SII), 3 |
| chr10_-_3827417 | 0.09 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chr6_+_152128371 | 0.09 |
ENST00000443427.1
|
ESR1
|
estrogen receptor 1 |
| chr14_-_57277163 | 0.09 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr1_+_156211753 | 0.09 |
ENST00000368272.4
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 0.5 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.2 | GO:0090260 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.5 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.4 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:1903799 | prostate epithelial cord elongation(GO:0060523) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0033594 | response to vitamin K(GO:0032571) response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 1.2 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0033292 | T-tubule organization(GO:0033292) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.2 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.1 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |