Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
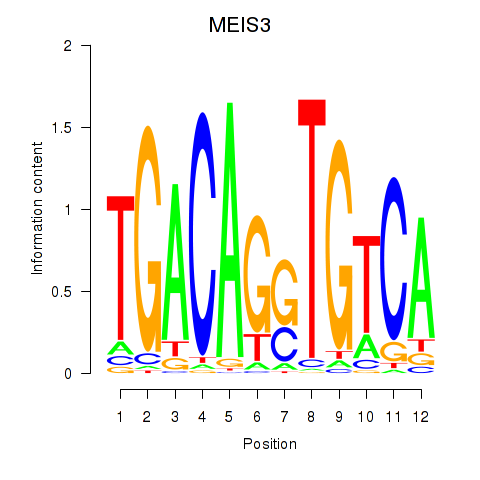
Results for MEIS3_TGIF2LX
Z-value: 0.48
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2LX | hg19_v2_chrX_+_89176881_89176955 | -0.19 | 3.0e-01 | Click! |
| MEIS3 | hg19_v2_chr19_-_47922750_47922795 | -0.05 | 8.0e-01 | Click! |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_32557610 | 1.35 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr11_+_86511549 | 1.16 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr1_-_47655686 | 0.91 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr19_+_17982747 | 0.88 |
ENST00000222248.3
|
SLC5A5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr19_-_9092018 | 0.83 |
ENST00000397910.4
|
MUC16
|
mucin 16, cell surface associated |
| chr1_+_79115503 | 0.70 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr11_+_827553 | 0.56 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr6_+_24495067 | 0.51 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr3_-_122512619 | 0.51 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr6_-_52860171 | 0.49 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr19_-_45826125 | 0.48 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr13_+_96085847 | 0.47 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr16_+_30212378 | 0.44 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_+_48172639 | 0.44 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr2_-_73460334 | 0.43 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr11_-_82745238 | 0.42 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr16_+_69458428 | 0.42 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr18_-_52989525 | 0.39 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_-_3221358 | 0.38 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr1_-_155162658 | 0.37 |
ENST00000368389.2
ENST00000368396.4 ENST00000343256.5 ENST00000342482.4 ENST00000368398.3 ENST00000368390.3 ENST00000337604.5 ENST00000368392.3 ENST00000438413.1 ENST00000368393.3 ENST00000457295.2 ENST00000338684.5 ENST00000368395.1 |
MUC1
|
mucin 1, cell surface associated |
| chr7_+_5919458 | 0.37 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr17_-_71088797 | 0.37 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr11_+_86511569 | 0.36 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr11_+_102188272 | 0.36 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr11_+_102188224 | 0.34 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr16_+_1306060 | 0.33 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chr7_+_134331550 | 0.33 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr9_+_96026230 | 0.32 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr12_-_81763127 | 0.31 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_+_24714729 | 0.31 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr18_+_3449330 | 0.31 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_+_97202480 | 0.30 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr5_+_125695805 | 0.30 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_81763184 | 0.29 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_+_121647962 | 0.29 |
ENST00000542067.1
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr6_-_52859968 | 0.28 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_+_159750720 | 0.27 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr2_+_97203082 | 0.27 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr1_+_159750776 | 0.27 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chrX_+_148622513 | 0.25 |
ENST00000393985.3
ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr13_-_67802549 | 0.24 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr17_-_19648916 | 0.24 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_+_57232690 | 0.23 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr17_+_34538310 | 0.23 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr1_+_161736072 | 0.22 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr10_+_114169299 | 0.22 |
ENST00000369410.3
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr3_+_9691117 | 0.22 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr3_+_52719936 | 0.22 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr18_+_76829385 | 0.22 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr3_+_122513642 | 0.22 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr3_-_49726486 | 0.22 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_+_61447845 | 0.22 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr21_-_43735446 | 0.21 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr1_+_11333245 | 0.20 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr17_+_28256874 | 0.20 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr17_-_29233769 | 0.20 |
ENST00000581216.1
|
TEFM
|
transcription elongation factor, mitochondrial |
| chr17_+_18380051 | 0.19 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr12_+_121647868 | 0.18 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr16_+_29789561 | 0.18 |
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16 |
| chr15_-_78591912 | 0.18 |
ENST00000560569.1
ENST00000558459.1 ENST00000558311.1 |
WDR61
|
WD repeat domain 61 |
| chr19_-_51531210 | 0.18 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr15_-_78592053 | 0.17 |
ENST00000267973.2
|
WDR61
|
WD repeat domain 61 |
| chr11_-_76381029 | 0.17 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr19_-_4559814 | 0.17 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr2_-_55920952 | 0.17 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr16_-_69418649 | 0.17 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chrX_-_13835147 | 0.16 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_140947563 | 0.16 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr5_+_122110691 | 0.16 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr22_-_50050986 | 0.16 |
ENST00000405854.1
|
C22orf34
|
chromosome 22 open reading frame 34 |
| chr3_-_52443799 | 0.16 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr11_+_63304273 | 0.16 |
ENST00000439013.2
ENST00000255688.3 |
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3 |
| chr9_-_86432547 | 0.16 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr19_-_51531272 | 0.16 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr19_+_12273866 | 0.15 |
ENST00000425827.1
ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr3_-_52719810 | 0.15 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr15_-_74284613 | 0.15 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr19_-_51530916 | 0.15 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr22_-_50051151 | 0.15 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr9_-_99540328 | 0.15 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr11_-_59612969 | 0.14 |
ENST00000541311.1
ENST00000257248.2 |
GIF
|
gastric intrinsic factor (vitamin B synthesis) |
| chr16_+_1290694 | 0.14 |
ENST00000338844.3
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr2_+_149402553 | 0.14 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_+_10560147 | 0.14 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr16_+_1290725 | 0.14 |
ENST00000461509.2
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr12_+_104458235 | 0.14 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr3_-_130745571 | 0.13 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr2_+_38177575 | 0.13 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr8_-_98290087 | 0.13 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr11_-_76381781 | 0.13 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr8_+_7345191 | 0.13 |
ENST00000335510.6
|
DEFB105B
|
defensin, beta 105B |
| chr17_+_33448593 | 0.13 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr2_+_220379052 | 0.13 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr8_-_7681411 | 0.13 |
ENST00000334773.6
|
DEFB105A
|
defensin, beta 105A |
| chr19_-_51529849 | 0.13 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr13_+_33160553 | 0.13 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr7_+_116654935 | 0.12 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr12_+_113682066 | 0.12 |
ENST00000392569.4
ENST00000552542.1 |
TPCN1
|
two pore segment channel 1 |
| chr15_+_85427903 | 0.12 |
ENST00000286749.3
ENST00000394573.1 ENST00000537703.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr19_+_35773242 | 0.11 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr2_-_179914760 | 0.11 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr11_+_18287721 | 0.11 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr4_+_175839506 | 0.11 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chrX_-_23926004 | 0.11 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr11_+_18287801 | 0.11 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr17_-_76220740 | 0.10 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr10_+_104535994 | 0.10 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr6_+_24495185 | 0.10 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr11_-_130786333 | 0.10 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr1_+_154975258 | 0.10 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr17_+_40985407 | 0.10 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr22_-_31885514 | 0.10 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr4_+_175839551 | 0.10 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr9_-_130541017 | 0.10 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr1_+_210406121 | 0.10 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr16_-_20364030 | 0.10 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr7_+_76139833 | 0.10 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr10_+_104503727 | 0.10 |
ENST00000448841.1
|
WBP1L
|
WW domain binding protein 1-like |
| chr7_-_144100786 | 0.10 |
ENST00000223140.5
|
NOBOX
|
NOBOX oogenesis homeobox |
| chr16_-_20364122 | 0.10 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr4_+_159727272 | 0.10 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr3_+_29322437 | 0.10 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr8_-_7243080 | 0.10 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr11_+_117947724 | 0.10 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr11_+_117947782 | 0.10 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_+_100598691 | 0.10 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr17_-_28257080 | 0.09 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr20_-_3644046 | 0.09 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr16_-_2004683 | 0.09 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr1_-_54872059 | 0.09 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr6_+_131148538 | 0.09 |
ENST00000541421.2
|
SMLR1
|
small leucine-rich protein 1 |
| chr11_-_18956556 | 0.09 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chrX_+_148622138 | 0.08 |
ENST00000450602.2
ENST00000441248.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr16_+_72090053 | 0.08 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr19_+_5455421 | 0.08 |
ENST00000222033.4
|
ZNRF4
|
zinc and ring finger 4 |
| chr1_+_154975110 | 0.08 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr7_+_76139741 | 0.08 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr6_+_50681541 | 0.08 |
ENST00000008391.3
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr2_-_198299726 | 0.08 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr17_-_5015129 | 0.08 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr20_-_57089934 | 0.08 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr11_-_111749878 | 0.08 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr17_+_58755184 | 0.08 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr18_+_54318566 | 0.08 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr11_-_111749767 | 0.08 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr12_+_52203789 | 0.07 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr6_+_42883727 | 0.07 |
ENST00000304672.1
ENST00000441198.1 ENST00000446507.1 |
PTCRA
|
pre T-cell antigen receptor alpha |
| chr4_+_78783674 | 0.07 |
ENST00000315567.8
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr12_+_100867486 | 0.07 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr3_-_47023455 | 0.07 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr16_+_1306093 | 0.07 |
ENST00000211076.3
|
TPSD1
|
tryptase delta 1 |
| chr16_-_18468926 | 0.07 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr17_+_34431212 | 0.07 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr9_+_131843377 | 0.07 |
ENST00000372546.4
ENST00000406974.3 ENST00000540102.1 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr1_+_19923454 | 0.07 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr10_+_51187938 | 0.07 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr10_-_7661623 | 0.06 |
ENST00000298441.6
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr12_-_56615693 | 0.06 |
ENST00000394013.2
ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41
|
ring finger protein 41 |
| chr10_-_31320860 | 0.06 |
ENST00000436087.2
ENST00000442986.1 ENST00000413025.1 ENST00000452305.1 |
ZNF438
|
zinc finger protein 438 |
| chr3_+_147795932 | 0.06 |
ENST00000490465.1
|
RP11-639B1.1
|
RP11-639B1.1 |
| chr15_+_83478370 | 0.06 |
ENST00000286760.4
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr22_+_21369316 | 0.06 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr1_+_158325684 | 0.05 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chrX_-_117250740 | 0.05 |
ENST00000371882.1
ENST00000540167.1 ENST00000545703.1 |
KLHL13
|
kelch-like family member 13 |
| chr12_+_120875910 | 0.05 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr19_+_11909329 | 0.05 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr11_+_45868957 | 0.05 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr17_+_7210294 | 0.05 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_+_219524473 | 0.05 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr7_+_76139925 | 0.05 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr9_-_37592561 | 0.05 |
ENST00000544379.1
ENST00000377773.5 ENST00000401811.3 ENST00000321301.6 |
TOMM5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr4_-_40517984 | 0.05 |
ENST00000381795.6
|
RBM47
|
RNA binding motif protein 47 |
| chr2_-_89597542 | 0.05 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr1_-_201081579 | 0.05 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr3_+_49726932 | 0.04 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr2_+_233271546 | 0.04 |
ENST00000295453.3
|
ALPPL2
|
alkaline phosphatase, placental-like 2 |
| chr10_-_105212141 | 0.04 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr15_-_51914996 | 0.04 |
ENST00000251076.5
|
DMXL2
|
Dmx-like 2 |
| chr8_-_126104055 | 0.04 |
ENST00000318410.7
|
KIAA0196
|
KIAA0196 |
| chrX_-_13835461 | 0.04 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr11_+_67776012 | 0.04 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr8_-_126103969 | 0.04 |
ENST00000517845.1
|
KIAA0196
|
KIAA0196 |
| chr11_+_111749650 | 0.04 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr8_+_99439214 | 0.04 |
ENST00000287042.4
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr18_+_3449695 | 0.04 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_156803329 | 0.04 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr2_+_162087577 | 0.04 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_11445856 | 0.04 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr11_+_64073699 | 0.04 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr7_-_105332084 | 0.03 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr7_-_48068643 | 0.03 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr11_+_19138670 | 0.03 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr18_+_3449821 | 0.03 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_133748913 | 0.03 |
ENST00000310926.4
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr2_+_27799389 | 0.03 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr1_-_116383738 | 0.03 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr7_-_48068671 | 0.03 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr11_-_61348292 | 0.03 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chrX_+_150863596 | 0.03 |
ENST00000448726.1
ENST00000538575.1 |
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr4_+_671711 | 0.03 |
ENST00000400159.2
|
MYL5
|
myosin, light chain 5, regulatory |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.6 | GO:0006540 | acetate metabolic process(GO:0006083) glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.7 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.2 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.5 | GO:0032308 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.4 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0034255 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 1.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |