Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
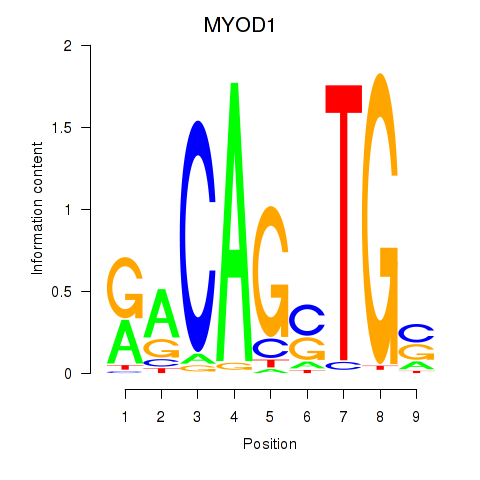
Results for MYOD1
Z-value: 0.85
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg19_v2_chr11_+_17741111_17741124 | -0.01 | 9.5e-01 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_67427389 | 5.49 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_-_13043697 | 3.75 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr3_-_50383096 | 3.37 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr16_+_67465016 | 2.98 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr11_+_27062860 | 2.97 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_68165657 | 2.49 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_85981008 | 2.44 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr7_-_120497178 | 2.17 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr11_+_86511549 | 1.99 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chrX_-_99665262 | 1.97 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr13_-_26625169 | 1.90 |
ENST00000319420.3
|
SHISA2
|
shisa family member 2 |
| chr5_-_150603679 | 1.85 |
ENST00000355417.2
|
CCDC69
|
coiled-coil domain containing 69 |
| chr12_+_50355647 | 1.83 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr3_+_111393659 | 1.77 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_-_154335300 | 1.77 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr6_-_24911195 | 1.77 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr16_-_84273304 | 1.77 |
ENST00000308251.4
ENST00000568181.1 |
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr17_-_46035187 | 1.74 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr11_+_27062272 | 1.70 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_+_165675269 | 1.68 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr11_+_27062502 | 1.60 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_162351503 | 1.56 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr19_-_7990991 | 1.50 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr7_-_73184588 | 1.46 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr22_-_36013368 | 1.42 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr10_+_94608245 | 1.38 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr1_+_87794150 | 1.30 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr6_+_17281573 | 1.29 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr2_-_220435963 | 1.24 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr1_+_60280458 | 1.21 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr12_+_56473628 | 1.20 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr9_+_36136700 | 1.17 |
ENST00000396613.3
ENST00000377959.1 ENST00000377960.4 |
GLIPR2
|
GLI pathogenesis-related 2 |
| chr1_+_89990431 | 1.17 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr4_-_23891693 | 1.16 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr5_-_159739532 | 1.16 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr9_-_80263220 | 1.15 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr14_-_100625932 | 1.14 |
ENST00000553834.1
|
DEGS2
|
delta(4)-desaturase, sphingolipid 2 |
| chr3_-_66024213 | 1.14 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_+_45918092 | 1.11 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr2_+_223289208 | 1.08 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr7_+_73442487 | 1.07 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr5_+_131630117 | 1.04 |
ENST00000200652.3
|
SLC22A4
|
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
| chr11_-_33891362 | 1.03 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_223537475 | 1.01 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr2_+_121103706 | 1.00 |
ENST00000295228.3
|
INHBB
|
inhibin, beta B |
| chr1_-_154928562 | 1.00 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr11_+_6411670 | 0.98 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr16_+_83932684 | 0.98 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr1_-_226129083 | 0.96 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr8_-_121824374 | 0.96 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr20_+_58179582 | 0.95 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr11_+_6411636 | 0.95 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr15_-_42186248 | 0.94 |
ENST00000320955.6
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5 |
| chr1_-_40367668 | 0.94 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_-_226129189 | 0.93 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr5_+_50678921 | 0.91 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr1_-_223537401 | 0.91 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr21_-_34185944 | 0.90 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr7_+_73442422 | 0.89 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr17_-_72968809 | 0.89 |
ENST00000530857.1
ENST00000425042.2 |
HID1
|
HID1 domain containing |
| chr11_-_108464465 | 0.88 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr5_+_169532896 | 0.87 |
ENST00000306268.6
ENST00000449804.2 |
FOXI1
|
forkhead box I1 |
| chr6_+_149068464 | 0.86 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr7_+_107301065 | 0.86 |
ENST00000265715.3
|
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr4_+_129732467 | 0.86 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr1_-_57285038 | 0.84 |
ENST00000343433.6
|
C1orf168
|
chromosome 1 open reading frame 168 |
| chr17_-_7216939 | 0.82 |
ENST00000573684.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr1_-_75139397 | 0.82 |
ENST00000326665.5
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr6_-_33714752 | 0.81 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr11_+_10476851 | 0.81 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr1_+_3614591 | 0.80 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr12_+_108525517 | 0.79 |
ENST00000332082.4
|
WSCD2
|
WSC domain containing 2 |
| chr17_+_76165213 | 0.79 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr11_+_86511569 | 0.78 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr12_+_122150646 | 0.77 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr7_+_73442102 | 0.76 |
ENST00000445912.1
ENST00000252034.7 |
ELN
|
elastin |
| chr11_-_108464321 | 0.76 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr2_+_95537170 | 0.76 |
ENST00000295201.4
|
TEKT4
|
tektin 4 |
| chr18_-_54305658 | 0.75 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr5_+_139027877 | 0.73 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr3_-_121740969 | 0.73 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr19_-_47137942 | 0.72 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr7_+_116660246 | 0.72 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr8_-_144691718 | 0.72 |
ENST00000377579.3
ENST00000433751.1 ENST00000220966.6 |
PYCRL
|
pyrroline-5-carboxylate reductase-like |
| chr8_-_29120580 | 0.71 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr1_+_159750720 | 0.69 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr1_+_159750776 | 0.68 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr17_+_4981535 | 0.68 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr5_+_102201687 | 0.68 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_233562015 | 0.67 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr3_+_108308513 | 0.67 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr2_+_24714729 | 0.67 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr5_+_102201722 | 0.66 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_62584197 | 0.66 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr1_-_204329013 | 0.65 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr1_+_61869748 | 0.65 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr16_+_88772866 | 0.64 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chrX_+_150151752 | 0.64 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr8_-_130951940 | 0.62 |
ENST00000522250.1
ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr11_-_2292182 | 0.62 |
ENST00000331289.4
|
ASCL2
|
achaete-scute family bHLH transcription factor 2 |
| chr5_+_102201509 | 0.61 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_89299885 | 0.61 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr7_+_73442457 | 0.61 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr17_-_43339474 | 0.61 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr2_-_220436248 | 0.60 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr5_+_139028510 | 0.60 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr17_-_43339453 | 0.59 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr14_+_22739823 | 0.59 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr14_-_24911448 | 0.59 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr17_+_26369865 | 0.58 |
ENST00000582037.1
|
NLK
|
nemo-like kinase |
| chr7_+_150065278 | 0.58 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr17_+_76164639 | 0.58 |
ENST00000225777.3
ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2
|
synaptogyrin 2 |
| chr8_-_17270809 | 0.58 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr4_+_166794862 | 0.57 |
ENST00000513213.1
|
TLL1
|
tolloid-like 1 |
| chr19_+_35521572 | 0.57 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr11_-_73694346 | 0.56 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr4_+_89299994 | 0.56 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr17_-_47755436 | 0.56 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr6_+_133562472 | 0.55 |
ENST00000430974.2
ENST00000367895.5 ENST00000355167.3 ENST00000355286.6 |
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr6_+_138483058 | 0.55 |
ENST00000251691.4
|
KIAA1244
|
KIAA1244 |
| chr8_+_81397876 | 0.54 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chrX_-_108868390 | 0.53 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr17_-_42277203 | 0.53 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr14_-_24912047 | 0.53 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr3_+_32147997 | 0.53 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr5_+_56111361 | 0.53 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr1_+_36023370 | 0.53 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr11_+_71938925 | 0.52 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr18_-_53089723 | 0.52 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr17_+_33474826 | 0.52 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr17_+_33474860 | 0.52 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr15_+_97326659 | 0.52 |
ENST00000558553.1
|
SPATA8
|
spermatogenesis associated 8 |
| chr11_-_88070920 | 0.52 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr7_+_35756186 | 0.51 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr1_-_167905225 | 0.51 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr8_+_81398444 | 0.51 |
ENST00000455036.3
ENST00000426744.2 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr16_-_31076332 | 0.50 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr8_-_91657740 | 0.50 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr3_-_65583561 | 0.49 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_+_113970772 | 0.49 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr9_+_136399929 | 0.49 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr11_-_8954491 | 0.49 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr10_-_73848531 | 0.49 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_+_16062820 | 0.48 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chrX_+_115567767 | 0.48 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr1_-_86043921 | 0.47 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr8_+_67341239 | 0.47 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chrX_-_99986494 | 0.47 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr16_+_58059470 | 0.47 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr1_-_185286461 | 0.46 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr17_+_40985407 | 0.46 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr5_+_102201430 | 0.46 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_-_62149433 | 0.46 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_+_11060004 | 0.46 |
ENST00000542579.1
ENST00000399850.3 ENST00000417956.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr19_+_3094398 | 0.46 |
ENST00000078429.4
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
| chr16_+_71660079 | 0.45 |
ENST00000565261.1
ENST00000268485.3 ENST00000299952.4 |
MARVELD3
|
MARVEL domain containing 3 |
| chr16_-_88772670 | 0.45 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr2_+_95963052 | 0.45 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr12_-_117628253 | 0.44 |
ENST00000330622.5
|
FBXO21
|
F-box protein 21 |
| chr19_+_14544099 | 0.44 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr16_+_103816 | 0.44 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr9_-_128003606 | 0.43 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr6_+_44184653 | 0.43 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr8_-_91657909 | 0.43 |
ENST00000418210.2
|
TMEM64
|
transmembrane protein 64 |
| chr22_+_17082732 | 0.43 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr3_-_49851313 | 0.43 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr2_-_178417742 | 0.43 |
ENST00000408939.3
|
TTC30B
|
tetratricopeptide repeat domain 30B |
| chr16_-_12009735 | 0.43 |
ENST00000439887.2
ENST00000434724.2 |
GSPT1
|
G1 to S phase transition 1 |
| chr1_-_33647267 | 0.43 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr11_+_7597639 | 0.43 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_+_40839369 | 0.42 |
ENST00000372718.3
|
SMAP2
|
small ArfGAP2 |
| chr2_-_227050079 | 0.42 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr18_-_52989525 | 0.42 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr11_-_67275542 | 0.42 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr8_-_72268968 | 0.42 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr7_-_150675372 | 0.42 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_-_73937244 | 0.41 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr8_-_37594944 | 0.41 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr15_-_45670924 | 0.40 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr11_-_119252359 | 0.40 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr17_-_71088797 | 0.40 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr3_+_52454971 | 0.40 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr5_+_159343688 | 0.40 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr14_-_24551137 | 0.40 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr14_-_24551195 | 0.40 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr16_-_31076273 | 0.40 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr17_-_56032602 | 0.39 |
ENST00000577840.1
|
CUEDC1
|
CUE domain containing 1 |
| chr14_-_106453155 | 0.39 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr14_-_89021077 | 0.39 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr12_-_56122426 | 0.39 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr11_+_1942580 | 0.39 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr16_+_23847267 | 0.39 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr2_-_37899323 | 0.39 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr19_+_19030497 | 0.39 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr16_+_71660052 | 0.39 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr11_+_71238313 | 0.39 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr16_-_88772761 | 0.38 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr19_+_19030478 | 0.38 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr19_+_16771936 | 0.38 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr11_-_117698765 | 0.38 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 2.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.6 | 6.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 1.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 1.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 1.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.3 | 0.9 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.3 | 0.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.3 | 0.8 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.3 | 1.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.2 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 1.0 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 3.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.7 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.2 | 0.6 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 0.8 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 1.0 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 3.0 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.2 | 0.5 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.2 | 1.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 0.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 0.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.4 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.4 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.8 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.5 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 2.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.7 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.6 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 1.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 0.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 3.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.5 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.9 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 1.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.9 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.1 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 2.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.5 | GO:0060482 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.1 | 1.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.6 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 5.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.3 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.6 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.3 | GO:0070940 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.5 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 3.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 2.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.7 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 1.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.3 | GO:0006915 | apoptotic process(GO:0006915) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.4 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.6 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.3 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 1.2 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.1 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 3.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 1.8 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.5 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 2.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 3.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.0 | 3.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 2.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 0.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 1.0 | GO:0008513 | secondary active organic cation transmembrane transporter activity(GO:0008513) |
| 0.2 | 0.7 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.8 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.2 | 0.7 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.6 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.3 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 2.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 3.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.5 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 2.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.0 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 5.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |