Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
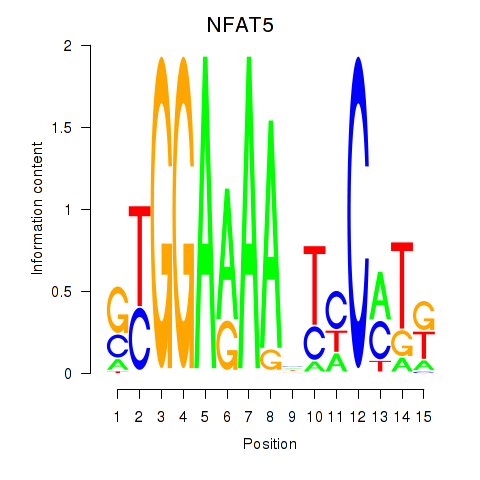
Results for NFAT5
Z-value: 0.75
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.16 | nuclear factor of activated T cells 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFAT5 | hg19_v2_chr16_+_69599861_69599887, hg19_v2_chr16_+_69600058_69600111 | 0.53 | 2.5e-03 | Click! |
Activity profile of NFAT5 motif
Sorted Z-values of NFAT5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_111888890 | 3.99 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr1_+_111889212 | 3.88 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr18_-_24765248 | 3.86 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr9_-_80263220 | 3.34 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr20_-_44176391 | 2.72 |
ENST00000409554.1
|
EPPIN
|
epididymal peptidase inhibitor |
| chr17_+_45908974 | 2.62 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr20_-_44176060 | 2.53 |
ENST00000354280.4
ENST00000504988.1 |
EPPIN
EPPIN-WFDC6
|
epididymal peptidase inhibitor EPPIN-WFDC6 readthrough |
| chr17_+_68071458 | 2.53 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_68071389 | 2.51 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_+_156712372 | 2.26 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_60539405 | 2.24 |
ENST00000450089.2
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_-_60539422 | 2.20 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr8_-_133772794 | 2.05 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr5_+_140186647 | 1.83 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr12_+_69742121 | 1.82 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr1_+_54359854 | 1.80 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr20_-_44176013 | 1.77 |
ENST00000555685.1
|
EPPIN
|
epididymal peptidase inhibitor |
| chr16_+_2079637 | 1.64 |
ENST00000561844.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr12_+_119772502 | 1.57 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr1_+_95975672 | 1.40 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr3_+_49027308 | 1.40 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr14_+_91580777 | 1.36 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr14_+_91580708 | 1.35 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr14_+_91581011 | 1.34 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr11_-_75062829 | 1.29 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr5_+_140261703 | 1.24 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr16_+_2079501 | 1.23 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr5_-_41261540 | 1.19 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr4_-_83483395 | 1.17 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr19_+_14142535 | 1.14 |
ENST00000263379.2
|
IL27RA
|
interleukin 27 receptor, alpha |
| chr13_-_24463530 | 1.12 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr4_+_74718906 | 1.09 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr6_+_153552455 | 1.08 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr4_+_17579110 | 1.06 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr14_+_100848311 | 1.06 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr14_-_102706934 | 1.06 |
ENST00000523231.1
ENST00000524370.1 ENST00000517966.1 |
MOK
|
MOK protein kinase |
| chr1_-_57045228 | 1.05 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr7_-_47579188 | 1.01 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr6_-_134495992 | 0.97 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_-_55375622 | 0.88 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr9_+_139921916 | 0.86 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr19_+_859425 | 0.85 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr18_-_52989525 | 0.84 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr11_-_75062730 | 0.77 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr16_+_30968615 | 0.76 |
ENST00000262519.8
|
SETD1A
|
SET domain containing 1A |
| chr8_-_27469196 | 0.75 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr14_+_91580732 | 0.75 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr3_+_69812877 | 0.73 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr2_+_56179262 | 0.70 |
ENST00000606639.1
|
RP11-481J13.1
|
RP11-481J13.1 |
| chr4_+_17578815 | 0.68 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr1_-_246670519 | 0.66 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr11_+_103907308 | 0.64 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr2_-_20101385 | 0.62 |
ENST00000431392.1
|
TTC32
|
tetratricopeptide repeat domain 32 |
| chr1_-_145470383 | 0.61 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr17_-_38083843 | 0.61 |
ENST00000304046.2
ENST00000579695.1 |
ORMDL3
|
ORM1-like 3 (S. cerevisiae) |
| chr14_+_91580357 | 0.61 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr9_-_13175823 | 0.59 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr15_+_45028753 | 0.57 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_+_1115056 | 0.57 |
ENST00000379288.3
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr14_+_95078714 | 0.55 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr2_+_198570081 | 0.55 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr9_-_124989804 | 0.54 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr18_-_52989217 | 0.52 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr3_-_42003479 | 0.51 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr1_-_85870177 | 0.50 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr22_-_44208070 | 0.49 |
ENST00000396231.2
|
EFCAB6
|
EF-hand calcium binding domain 6 |
| chr15_+_49170281 | 0.49 |
ENST00000560490.1
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr3_-_9834375 | 0.49 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr14_-_25479811 | 0.47 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chrX_+_70752917 | 0.46 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr15_+_43885252 | 0.45 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_-_202579 | 0.45 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr17_+_32646055 | 0.42 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chr3_+_8543533 | 0.41 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr15_+_45028520 | 0.40 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr3_+_185000729 | 0.40 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr15_+_43985084 | 0.40 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_-_118928543 | 0.39 |
ENST00000419373.2
|
RP11-501J20.2
|
RP11-501J20.2 |
| chr20_+_33104199 | 0.39 |
ENST00000357156.2
ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1
|
dynein, light chain, roadblock-type 1 |
| chr13_-_67804445 | 0.39 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr10_+_111967345 | 0.39 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr15_+_49170083 | 0.39 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr15_+_45028719 | 0.38 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr14_+_103589789 | 0.38 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr10_+_89419370 | 0.38 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr5_-_35230434 | 0.37 |
ENST00000504500.1
|
PRLR
|
prolactin receptor |
| chr12_+_53848505 | 0.36 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr2_-_68479614 | 0.36 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr11_+_65190245 | 0.36 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr15_+_43985725 | 0.35 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_+_26322185 | 0.33 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_+_15852203 | 0.31 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr7_-_92157760 | 0.30 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr7_-_92157747 | 0.30 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr4_+_26322409 | 0.30 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_120502441 | 0.29 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr7_+_149570049 | 0.29 |
ENST00000421974.2
ENST00000456496.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr7_-_92855762 | 0.28 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr3_+_120461484 | 0.28 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr12_+_10365404 | 0.28 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr9_-_14314518 | 0.27 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr3_+_63805017 | 0.26 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr12_-_122018859 | 0.25 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr3_+_47866490 | 0.25 |
ENST00000457607.1
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr3_-_49131013 | 0.24 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr12_-_323689 | 0.24 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr16_-_51185172 | 0.21 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr16_+_68573116 | 0.21 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr9_-_6015607 | 0.20 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr16_-_75498553 | 0.20 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr7_+_95401851 | 0.19 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr1_+_36789335 | 0.19 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr13_+_45694583 | 0.19 |
ENST00000340473.6
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa |
| chrX_-_45060135 | 0.19 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr22_+_24551765 | 0.18 |
ENST00000337989.7
|
CABIN1
|
calcineurin binding protein 1 |
| chrX_+_83116142 | 0.18 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr6_+_138188551 | 0.18 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr17_-_15466742 | 0.18 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr8_+_96037205 | 0.18 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr20_-_23807358 | 0.17 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr1_+_34632484 | 0.17 |
ENST00000373374.3
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr17_-_15466850 | 0.16 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr16_-_67514982 | 0.16 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr7_+_95401877 | 0.16 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr2_-_162931052 | 0.14 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr1_-_36945097 | 0.14 |
ENST00000331941.5
ENST00000418048.2 ENST00000338937.5 ENST00000440588.2 |
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr11_+_19798964 | 0.13 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_+_22696314 | 0.13 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr11_+_19799327 | 0.13 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr3_-_44552094 | 0.13 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr17_+_40950797 | 0.12 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr17_+_4853442 | 0.12 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr20_-_62103862 | 0.12 |
ENST00000344462.4
ENST00000357249.2 ENST00000359125.2 ENST00000360480.3 ENST00000370224.1 ENST00000344425.5 ENST00000354587.3 ENST00000359689.1 |
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2 |
| chr4_+_122685691 | 0.11 |
ENST00000424958.1
|
AC079341.1
|
Uncharacterized protein |
| chr1_+_18958008 | 0.11 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chrX_+_12993202 | 0.11 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr2_+_47799601 | 0.11 |
ENST00000601243.1
|
AC138655.1
|
CDNA: FLJ23120 fis, clone LNG07989; HCG1987724; Uncharacterized protein |
| chr12_+_69139886 | 0.11 |
ENST00000398004.2
|
SLC35E3
|
solute carrier family 35, member E3 |
| chr12_+_99038998 | 0.10 |
ENST00000359972.2
ENST00000357310.1 ENST00000339433.3 ENST00000333991.1 |
APAF1
|
apoptotic peptidase activating factor 1 |
| chr6_+_89790459 | 0.10 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_-_99954788 | 0.09 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr4_-_76861392 | 0.09 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr6_+_109416684 | 0.09 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr12_-_11214893 | 0.08 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr18_-_74839891 | 0.08 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr10_-_135122603 | 0.07 |
ENST00000368563.2
|
TUBGCP2
|
tubulin, gamma complex associated protein 2 |
| chr5_-_137090028 | 0.07 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr7_-_102119342 | 0.07 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr12_-_75603482 | 0.07 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr12_-_75603643 | 0.06 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr7_-_19184929 | 0.06 |
ENST00000275461.3
|
FERD3L
|
Fer3-like bHLH transcription factor |
| chr6_+_160327974 | 0.05 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr17_+_18684563 | 0.05 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr7_+_65670186 | 0.05 |
ENST00000304842.5
ENST00000442120.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr3_-_149293990 | 0.04 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr8_+_96037255 | 0.04 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_-_133747589 | 0.04 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr17_+_40950900 | 0.04 |
ENST00000588527.1
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr19_-_46285736 | 0.04 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chr6_+_44094627 | 0.03 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr15_-_102285007 | 0.02 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr3_-_156878540 | 0.02 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr1_-_153517473 | 0.02 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr13_-_50265570 | 0.02 |
ENST00000378270.5
ENST00000378284.2 ENST00000378272.5 ENST00000378268.1 ENST00000242827.6 ENST00000378282.5 |
EBPL
|
emopamil binding protein-like |
| chr7_-_100888313 | 0.02 |
ENST00000442303.1
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chrX_+_18725758 | 0.02 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr11_+_56949221 | 0.01 |
ENST00000497933.1
|
LRRC55
|
leucine rich repeat containing 55 |
| chr7_-_100888337 | 0.01 |
ENST00000223136.4
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr4_-_74847800 | 0.00 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFAT5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.3 | 3.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 1.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 2.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 0.9 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 0.6 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 0.6 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.2 | 0.6 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 2.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.8 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 5.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 5.4 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 3.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.5 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.1 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.8 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.3 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.2 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 1.4 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 1.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 2.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.7 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 8.0 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 5.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.2 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 7.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 4.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) invadopodium membrane(GO:0071438) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 7.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.7 | 2.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.6 | 3.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.3 | 1.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 0.9 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.3 | 5.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 8.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 3.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 1.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 6.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 5.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |