Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
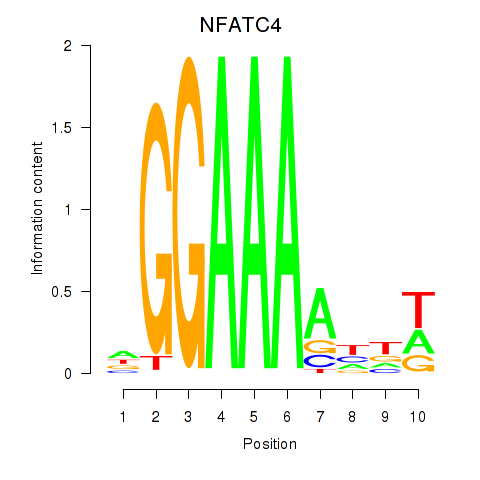
Results for NFATC4
Z-value: 1.09
Transcription factors associated with NFATC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC4
|
ENSG00000100968.9 | nuclear factor of activated T cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC4 | hg19_v2_chr14_+_24837226_24837547 | -0.42 | 2.1e-02 | Click! |
Activity profile of NFATC4 motif
Sorted Z-values of NFATC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_98703643 | 10.58 |
ENST00000477737.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr14_+_96949319 | 6.73 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr1_-_161337662 | 5.39 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr11_+_101918153 | 4.66 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr14_+_96858433 | 3.97 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr1_+_111889212 | 3.86 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr1_-_75139397 | 3.82 |
ENST00000326665.5
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr2_-_207629997 | 3.65 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr2_-_207630033 | 3.50 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr14_+_75536335 | 3.17 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_+_75536280 | 3.09 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr5_+_140254884 | 2.85 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr4_-_100356551 | 2.81 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr10_-_116444371 | 2.74 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr9_-_80263220 | 2.72 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr16_+_57769635 | 2.68 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr11_-_62474803 | 2.44 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_71551652 | 2.44 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_-_159869912 | 2.39 |
ENST00000368099.4
|
CCDC19
|
coiled-coil domain containing 19 |
| chr2_+_27237615 | 2.39 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_-_41261540 | 2.37 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr11_+_1942580 | 2.34 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr21_-_34185944 | 2.30 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr4_-_100356844 | 2.19 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_-_43412418 | 2.09 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr12_-_58329888 | 1.97 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr4_-_100356291 | 1.96 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_+_53343577 | 1.95 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr22_-_39548627 | 1.95 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr12_+_51318513 | 1.89 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr1_+_212782012 | 1.74 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr6_-_152639479 | 1.73 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr8_-_110703819 | 1.72 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr14_+_21156915 | 1.66 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr11_+_6866883 | 1.64 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr12_-_49582978 | 1.59 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr8_+_94767109 | 1.57 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr11_+_36616355 | 1.57 |
ENST00000532470.2
|
C11orf74
|
chromosome 11 open reading frame 74 |
| chr2_+_98330009 | 1.52 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr5_-_124081008 | 1.43 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr10_+_81838411 | 1.42 |
ENST00000372281.3
ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254
|
transmembrane protein 254 |
| chr17_-_5026397 | 1.41 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chrX_+_36254051 | 1.40 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr10_-_62704005 | 1.38 |
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr3_-_114035026 | 1.35 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr14_-_92413353 | 1.32 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr17_-_39165366 | 1.24 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr7_-_105319536 | 1.21 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr15_-_52587945 | 1.20 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr3_+_160559931 | 1.18 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr1_+_61869748 | 1.17 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr15_-_77712477 | 1.15 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr14_+_32546274 | 1.11 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr8_-_72274095 | 1.11 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr8_+_86157699 | 1.11 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr3_+_108321623 | 1.10 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr4_-_100212132 | 1.08 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr16_-_54963026 | 1.05 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chrX_+_22056165 | 1.04 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr17_-_39280419 | 1.03 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr2_-_242089677 | 1.02 |
ENST00000405260.1
|
PASK
|
PAS domain containing serine/threonine kinase |
| chr14_+_75988768 | 1.00 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr10_+_81838792 | 1.00 |
ENST00000372273.3
|
TMEM254
|
transmembrane protein 254 |
| chr7_-_99679324 | 0.99 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr17_+_76311791 | 0.99 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr3_-_119379427 | 0.99 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr11_+_100862811 | 0.97 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr14_+_62585332 | 0.97 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr12_-_39734783 | 0.96 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr9_+_92219919 | 0.95 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_-_149051194 | 0.92 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr1_+_79115503 | 0.91 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr5_-_135231516 | 0.91 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
| chr4_-_107957454 | 0.90 |
ENST00000285311.3
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr8_-_57123815 | 0.90 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr3_+_184055240 | 0.90 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr14_+_58754751 | 0.89 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr6_+_122720681 | 0.88 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr14_-_89883412 | 0.88 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr1_+_61547894 | 0.85 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr14_+_81421710 | 0.84 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr6_-_134495992 | 0.84 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_-_42811986 | 0.83 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_-_46769694 | 0.83 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr1_-_160231451 | 0.82 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr6_+_127898312 | 0.82 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_-_57045228 | 0.82 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr6_+_6588902 | 0.81 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr1_+_164528866 | 0.81 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr3_-_114790179 | 0.81 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_-_96076334 | 0.80 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr9_-_13165457 | 0.80 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr6_-_154677900 | 0.80 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_-_121740969 | 0.79 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr12_-_95945246 | 0.78 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr14_+_88852059 | 0.77 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr5_+_43602750 | 0.77 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr14_+_81421921 | 0.74 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr8_+_28747884 | 0.74 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr6_+_90272027 | 0.73 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr11_+_93063878 | 0.73 |
ENST00000298050.3
|
CCDC67
|
coiled-coil domain containing 67 |
| chr21_-_34185989 | 0.73 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_-_47600320 | 0.73 |
ENST00000525720.1
ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr3_+_119316721 | 0.72 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr9_+_26956371 | 0.72 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr11_-_47600549 | 0.72 |
ENST00000430070.2
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr19_+_39759154 | 0.71 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr5_+_132009675 | 0.71 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr4_-_84256024 | 0.71 |
ENST00000311412.5
|
HPSE
|
heparanase |
| chr1_-_145470383 | 0.71 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr16_-_86588627 | 0.70 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chrX_+_9431324 | 0.70 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr7_-_120498357 | 0.70 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr5_-_42812143 | 0.69 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr10_+_118350468 | 0.69 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr5_+_43603229 | 0.69 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr6_-_15586238 | 0.68 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr14_-_21979428 | 0.67 |
ENST00000538267.1
ENST00000298717.4 |
METTL3
|
methyltransferase like 3 |
| chr4_+_26322185 | 0.67 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_3600356 | 0.66 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr4_-_1670632 | 0.66 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr12_+_109577202 | 0.66 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr3_+_69812877 | 0.65 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_46142505 | 0.64 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr12_-_49582593 | 0.64 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_-_90049828 | 0.64 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_90049878 | 0.64 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr11_-_62457371 | 0.63 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr14_+_75988851 | 0.63 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr17_+_79953310 | 0.61 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_-_116662593 | 0.61 |
ENST00000227665.4
|
APOA5
|
apolipoprotein A-V |
| chr12_-_9268707 | 0.61 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr2_-_101767715 | 0.61 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr17_+_48610074 | 0.59 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr3_+_119316689 | 0.59 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr16_+_67381263 | 0.58 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr16_+_78056412 | 0.58 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr7_-_64023441 | 0.58 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr14_+_22636283 | 0.57 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chrX_+_15518923 | 0.57 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_-_151299842 | 0.57 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr6_+_140175987 | 0.56 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr11_-_46142615 | 0.56 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr10_+_97733786 | 0.56 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr1_-_151431909 | 0.56 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chrX_+_76709648 | 0.55 |
ENST00000439435.1
|
FGF16
|
fibroblast growth factor 16 |
| chr18_+_32556892 | 0.55 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_174846570 | 0.55 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_26322409 | 0.54 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_186560462 | 0.53 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560476 | 0.52 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr5_+_167718604 | 0.52 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr11_-_46142948 | 0.52 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr16_+_67381289 | 0.52 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr11_+_66234216 | 0.52 |
ENST00000349459.6
ENST00000320740.7 ENST00000524466.1 ENST00000526296.1 |
PELI3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr19_-_10697895 | 0.52 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chrX_-_134478012 | 0.51 |
ENST00000370766.3
|
ZNF75D
|
zinc finger protein 75D |
| chr5_+_149109825 | 0.50 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chrX_-_84634708 | 0.50 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr2_-_152830479 | 0.50 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chrX_-_65259914 | 0.50 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr2_+_233562015 | 0.50 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr12_-_89918522 | 0.49 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chrX_-_65259900 | 0.48 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr11_-_10315741 | 0.47 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr7_+_142364193 | 0.47 |
ENST00000390397.2
|
TRBV24-1
|
T cell receptor beta variable 24-1 |
| chr9_+_104161123 | 0.47 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr2_+_86668464 | 0.47 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr3_+_138340067 | 0.47 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_36789335 | 0.46 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr7_-_25702669 | 0.46 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr20_+_15177480 | 0.46 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr17_+_7792101 | 0.46 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr6_-_110736742 | 0.45 |
ENST00000368924.3
ENST00000368923.3 |
DDO
|
D-aspartate oxidase |
| chrX_-_84634737 | 0.44 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr2_-_175712270 | 0.44 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr9_+_91933407 | 0.44 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chrY_+_20708557 | 0.43 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr7_-_141673573 | 0.43 |
ENST00000547270.1
|
TAS2R38
|
taste receptor, type 2, member 38 |
| chr7_+_120628731 | 0.43 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_88471468 | 0.43 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr20_-_30310693 | 0.42 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr1_-_23521222 | 0.42 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr3_-_93747425 | 0.42 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr4_+_81951957 | 0.41 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr5_+_155753745 | 0.40 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr3_-_27763803 | 0.40 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr1_+_163038565 | 0.40 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chrX_-_15619076 | 0.40 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr12_-_100656134 | 0.39 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr1_-_85870177 | 0.39 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr6_-_26056695 | 0.39 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr19_-_42348692 | 0.39 |
ENST00000330743.3
ENST00000601246.1 |
LYPD4
|
LY6/PLAUR domain containing 4 |
| chrX_+_10124977 | 0.39 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr15_+_43885252 | 0.39 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr6_+_123100853 | 0.38 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr3_+_148545586 | 0.38 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr3_+_94657086 | 0.38 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr19_+_52772821 | 0.38 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr11_-_61735029 | 0.37 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.8 | 2.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.7 | 2.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.6 | 2.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.6 | 1.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.5 | 7.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 1.2 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.4 | 1.6 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.4 | 1.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.3 | 2.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.5 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 2.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.6 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 1.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.2 | 0.7 | GO:0071288 | T-helper 1 cell lineage commitment(GO:0002296) cellular response to mercury ion(GO:0071288) |
| 0.2 | 0.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.2 | 1.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 9.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 0.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.6 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 1.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.8 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.6 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 2.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.4 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.2 | GO:0035922 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.6 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 2.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.6 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.8 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.3 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.9 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.7 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.7 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 2.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.9 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 2.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.7 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 1.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 1.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.4 | GO:0050778 | positive regulation of immune response(GO:0050778) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0070943 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.6 | GO:0090277 | positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 2.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 2.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 1.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 2.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 5.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 3.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 2.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 7.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 1.6 | 10.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.0 | 7.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.6 | 2.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.5 | 1.5 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 2.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 0.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 1.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 1.6 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 0.7 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.6 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 0.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 1.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 0.7 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.2 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.5 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 4.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0060230 | lipase binding(GO:0035473) lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 2.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 1.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 3.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 1.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 4.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 13.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |