Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
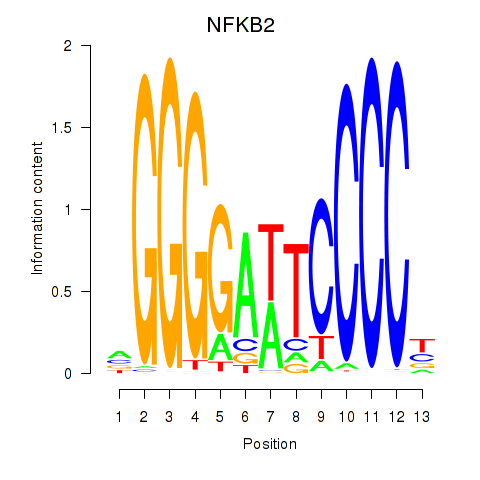
Results for NFKB2
Z-value: 0.60
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.13 | nuclear factor kappa B subunit 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg19_v2_chr10_+_104155450_104155479, hg19_v2_chr10_+_104154229_104154354 | 0.45 | 1.2e-02 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_74864386 | 1.58 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr1_-_186649543 | 1.29 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_7654057 | 1.19 |
ENST00000537121.1
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr22_-_37640456 | 1.10 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr22_-_37640277 | 1.06 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chrX_+_103173457 | 0.98 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr8_+_86376081 | 0.90 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr1_-_6545502 | 0.90 |
ENST00000535355.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr3_+_111718173 | 0.86 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr20_+_43803517 | 0.83 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr4_+_17812525 | 0.79 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr9_+_17579084 | 0.72 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr19_+_48824711 | 0.72 |
ENST00000599704.1
|
EMP3
|
epithelial membrane protein 3 |
| chr8_+_54793454 | 0.70 |
ENST00000276500.4
|
RGS20
|
regulator of G-protein signaling 20 |
| chr1_+_78354297 | 0.68 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr4_-_74904398 | 0.68 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr8_+_54793425 | 0.67 |
ENST00000522225.1
|
RGS20
|
regulator of G-protein signaling 20 |
| chr18_+_21529811 | 0.67 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chrX_+_64887512 | 0.66 |
ENST00000360270.5
|
MSN
|
moesin |
| chr1_-_38100539 | 0.66 |
ENST00000401069.1
|
RSPO1
|
R-spondin 1 |
| chr19_-_19051927 | 0.63 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr7_+_100547156 | 0.56 |
ENST00000379458.4
|
MUC3A
|
Protein LOC100131514 |
| chr5_-_137667459 | 0.55 |
ENST00000415130.2
ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C
|
cell division cycle 25C |
| chr19_-_19049791 | 0.54 |
ENST00000594439.1
ENST00000221222.11 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr6_+_32605195 | 0.53 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr16_-_74734672 | 0.53 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr19_-_41859814 | 0.53 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr3_-_50340996 | 0.52 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr16_-_74734742 | 0.52 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr8_-_62559366 | 0.51 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr3_+_111717600 | 0.51 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr1_-_47655686 | 0.51 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr21_-_45079341 | 0.50 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr3_+_111718036 | 0.49 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_-_38100491 | 0.49 |
ENST00000356545.2
|
RSPO1
|
R-spondin 1 |
| chr1_-_209824643 | 0.48 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr19_+_54371114 | 0.47 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr10_+_104154229 | 0.46 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr9_+_75263565 | 0.44 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr10_+_112257596 | 0.42 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr1_+_111770278 | 0.42 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr2_-_89310012 | 0.42 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr19_-_52227221 | 0.42 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chrX_-_15872914 | 0.41 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_64681641 | 0.41 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr1_-_159915386 | 0.40 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr1_+_111770232 | 0.40 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr7_-_28220354 | 0.40 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr8_+_90770008 | 0.40 |
ENST00000540020.1
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr7_+_120629653 | 0.39 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr4_-_74964904 | 0.39 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr15_-_89438742 | 0.39 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr8_-_145641864 | 0.38 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr11_-_18270182 | 0.38 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr5_+_60241020 | 0.37 |
ENST00000511107.1
ENST00000502658.1 |
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr16_+_88704978 | 0.37 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr19_+_45147098 | 0.37 |
ENST00000425690.3
ENST00000344956.4 ENST00000403059.4 |
PVR
|
poliovirus receptor |
| chr5_+_60240943 | 0.37 |
ENST00000296597.5
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr22_+_44319648 | 0.36 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr17_+_4854375 | 0.36 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr2_-_241737128 | 0.36 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr22_+_35796108 | 0.36 |
ENST00000382011.5
ENST00000416905.1 |
MCM5
|
minichromosome maintenance complex component 5 |
| chr19_-_50400212 | 0.36 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr3_-_108308241 | 0.36 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr20_+_60174827 | 0.35 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_+_150954493 | 0.35 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr10_+_124030819 | 0.35 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr1_-_231376836 | 0.34 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr19_-_4831701 | 0.34 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr22_+_35796056 | 0.34 |
ENST00000216122.4
|
MCM5
|
minichromosome maintenance complex component 5 |
| chr2_-_89292422 | 0.34 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chrX_+_68048803 | 0.33 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr11_-_76381781 | 0.33 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr19_+_45147313 | 0.33 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr8_+_95565947 | 0.32 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr2_+_173940668 | 0.32 |
ENST00000375213.3
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr16_+_15737124 | 0.32 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr22_+_44319619 | 0.32 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr19_+_10216899 | 0.31 |
ENST00000428358.1
ENST00000393796.4 ENST00000253107.7 ENST00000556468.1 ENST00000393793.1 |
PPAN-P2RY11
PPAN
|
PPAN-P2RY11 readthrough peter pan homolog (Drosophila) |
| chrX_+_150151824 | 0.31 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr8_-_145582118 | 0.31 |
ENST00000455319.2
ENST00000331890.5 |
FBXL6
|
F-box and leucine-rich repeat protein 6 |
| chr19_-_3786253 | 0.31 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr6_-_32157947 | 0.31 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr12_+_110011571 | 0.30 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr12_-_127256772 | 0.30 |
ENST00000536517.1
|
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr7_-_44229022 | 0.30 |
ENST00000403799.3
|
GCK
|
glucokinase (hexokinase 4) |
| chr1_-_208417620 | 0.30 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr2_-_89247338 | 0.30 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr15_-_75017711 | 0.30 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr17_-_39928106 | 0.30 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr20_-_43753104 | 0.30 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr6_+_32605134 | 0.30 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr19_+_30302805 | 0.29 |
ENST00000262643.3
ENST00000575243.1 ENST00000357943.5 |
CCNE1
|
cyclin E1 |
| chr9_-_139965000 | 0.29 |
ENST00000409687.3
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr8_+_26371763 | 0.29 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr14_+_97263641 | 0.29 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr19_-_3786354 | 0.29 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_150811705 | 0.28 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr14_-_51297197 | 0.28 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr9_+_38392661 | 0.28 |
ENST00000377698.3
|
ALDH1B1
|
aldehyde dehydrogenase 1 family, member B1 |
| chr18_+_268148 | 0.27 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr17_-_34207295 | 0.27 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr12_+_57522258 | 0.27 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr1_-_154943212 | 0.27 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr11_+_35639735 | 0.26 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr4_+_74702214 | 0.26 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr17_-_40333150 | 0.26 |
ENST00000264661.3
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr11_+_119056178 | 0.26 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr15_+_92937058 | 0.25 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr3_+_12329397 | 0.25 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr16_-_46864955 | 0.25 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_+_12329358 | 0.25 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr11_-_104840093 | 0.25 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr18_-_31802056 | 0.25 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr12_-_77272765 | 0.25 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr22_-_20255212 | 0.25 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr5_+_176730769 | 0.25 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr8_-_9008206 | 0.24 |
ENST00000310455.3
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr6_-_24646249 | 0.24 |
ENST00000430948.2
ENST00000537886.1 ENST00000535378.1 ENST00000378214.3 |
KIAA0319
|
KIAA0319 |
| chr20_+_35090150 | 0.24 |
ENST00000340491.4
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr7_+_142031986 | 0.24 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr5_+_176731572 | 0.24 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr1_-_28559502 | 0.24 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr9_-_123691047 | 0.24 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr16_-_68269971 | 0.23 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr6_-_43484621 | 0.23 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr10_+_13141585 | 0.23 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr9_-_71155783 | 0.23 |
ENST00000377311.3
|
TMEM252
|
transmembrane protein 252 |
| chr5_-_150466692 | 0.23 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_93426998 | 0.23 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr2_-_27718052 | 0.23 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr19_+_35485682 | 0.23 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr9_-_125667618 | 0.23 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr2_-_219157250 | 0.22 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chrX_-_15353629 | 0.22 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr2_+_173940163 | 0.22 |
ENST00000539448.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr20_-_48532019 | 0.22 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr6_-_43484718 | 0.22 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr1_-_154943002 | 0.22 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr10_-_86001210 | 0.22 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr12_+_8662057 | 0.22 |
ENST00000382064.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr20_-_42815733 | 0.21 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr16_-_28223166 | 0.21 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr19_-_50143452 | 0.21 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr1_-_183604794 | 0.21 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr19_+_19639670 | 0.21 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr17_+_38497640 | 0.21 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr16_+_31539197 | 0.21 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr8_+_32406179 | 0.21 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr17_-_27045405 | 0.21 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr17_-_5487277 | 0.21 |
ENST00000572272.1
ENST00000354411.3 ENST00000577119.1 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr7_-_111202511 | 0.20 |
ENST00000452895.1
ENST00000452753.1 ENST00000331762.3 |
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_+_31539183 | 0.20 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr1_+_901847 | 0.20 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr10_-_133795416 | 0.20 |
ENST00000540159.1
ENST00000368636.4 |
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr19_+_19639704 | 0.19 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr17_+_4855053 | 0.19 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr11_-_64570706 | 0.19 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_-_205744574 | 0.19 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr2_+_173940442 | 0.19 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr8_+_22857048 | 0.19 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr22_+_38349670 | 0.19 |
ENST00000442738.2
ENST00000460648.1 ENST00000407936.1 ENST00000488684.1 ENST00000492213.1 ENST00000606538.1 ENST00000405557.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr7_-_129691201 | 0.19 |
ENST00000480193.1
ENST00000360708.5 ENST00000311873.5 ENST00000481503.1 ENST00000358303.4 |
ZC3HC1
|
zinc finger, C3HC-type containing 1 |
| chr16_-_4465886 | 0.19 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr22_-_25801333 | 0.19 |
ENST00000444995.3
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like |
| chr8_-_37797621 | 0.18 |
ENST00000524298.1
ENST00000307599.4 |
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr1_-_6546001 | 0.18 |
ENST00000400913.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr7_-_44179972 | 0.18 |
ENST00000446581.1
|
MYL7
|
myosin, light chain 7, regulatory |
| chr2_-_70780770 | 0.18 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr21_+_17443521 | 0.18 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_31119615 | 0.18 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr17_-_27045427 | 0.18 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr1_-_205744205 | 0.18 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr8_+_128427857 | 0.18 |
ENST00000391675.1
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr19_+_1450112 | 0.18 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr11_+_706113 | 0.18 |
ENST00000318562.8
ENST00000533256.1 ENST00000534755.1 |
EPS8L2
|
EPS8-like 2 |
| chr17_-_42908155 | 0.18 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr12_-_49259643 | 0.18 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr3_+_113775576 | 0.18 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr21_+_17442799 | 0.18 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_70781087 | 0.18 |
ENST00000394241.3
ENST00000295400.6 |
TGFA
|
transforming growth factor, alpha |
| chr10_+_13142075 | 0.17 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr21_+_17443434 | 0.17 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_41328018 | 0.17 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr17_-_27045165 | 0.17 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr11_+_60691924 | 0.17 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr1_-_71513471 | 0.17 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chrX_-_141293047 | 0.17 |
ENST00000247452.3
|
MAGEC2
|
melanoma antigen family C, 2 |
| chr12_+_70760056 | 0.17 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr7_-_143059780 | 0.17 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr4_-_20985632 | 0.16 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr15_+_92397051 | 0.16 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr16_-_4466622 | 0.16 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr8_-_144923112 | 0.16 |
ENST00000442628.2
|
NRBP2
|
nuclear receptor binding protein 2 |
| chr15_+_92396920 | 0.16 |
ENST00000318445.6
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr9_+_115913222 | 0.16 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr14_+_21467414 | 0.16 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr7_-_151107767 | 0.16 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chr17_-_1389419 | 0.16 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr17_+_40172069 | 0.16 |
ENST00000393885.4
ENST00000393884.2 ENST00000587337.1 ENST00000479407.1 |
NKIRAS2
|
NFKB inhibitor interacting Ras-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 0.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.7 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 2.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 0.5 | GO:1900126 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 0.5 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.7 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.1 | 0.5 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 3.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.1 | GO:0045657 | positive regulation of macrophage differentiation(GO:0045651) positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.3 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.3 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.5 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.3 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.9 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.1 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:2000777 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 1.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 1.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.4 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 1.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.6 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 3.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.4 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 1.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.3 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.7 | GO:0036042 | lysophosphatidic acid binding(GO:0035727) long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 1.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |