Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
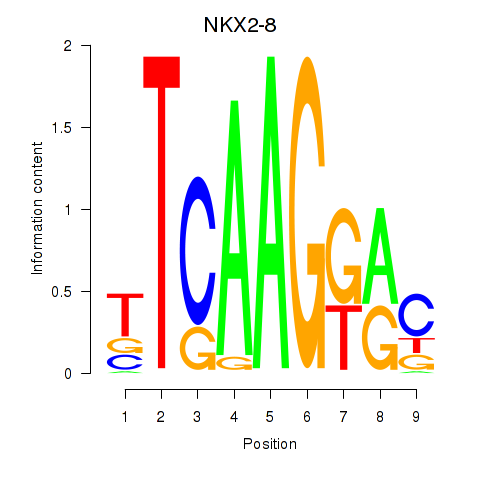
Results for NKX2-8
Z-value: 0.81
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | 0.53 | 2.5e-03 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13349650 | 3.12 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr1_+_120839412 | 2.70 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr15_+_40453204 | 2.29 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr17_+_76210367 | 2.15 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr1_+_152957707 | 2.01 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr14_-_24732403 | 1.87 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr19_-_51336443 | 1.65 |
ENST00000598673.1
|
KLK15
|
kallikrein-related peptidase 15 |
| chr17_+_76210267 | 1.64 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr8_+_27182862 | 1.54 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_+_44037546 | 1.53 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr14_-_24732368 | 1.49 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr1_+_154401791 | 1.44 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr20_+_58296265 | 1.44 |
ENST00000395636.2
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr20_+_31619454 | 1.28 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B, member 6 |
| chr2_+_136289030 | 0.99 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr11_-_128457446 | 0.97 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr11_-_107590383 | 0.93 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr15_-_70387120 | 0.87 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr2_+_101591314 | 0.87 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr3_-_53290016 | 0.85 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr3_-_47950745 | 0.83 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr14_-_106573756 | 0.81 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr1_+_10057274 | 0.79 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr5_-_78808617 | 0.78 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr13_-_23949671 | 0.77 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr1_-_94147385 | 0.77 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr16_+_8807419 | 0.76 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr16_+_88872176 | 0.76 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr8_-_62559366 | 0.69 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr17_-_8055747 | 0.69 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr6_+_69942298 | 0.69 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr2_-_36779411 | 0.67 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr11_+_35211429 | 0.66 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr14_-_106791536 | 0.66 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr12_-_118406777 | 0.65 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr7_+_135242652 | 0.64 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr22_-_50523760 | 0.63 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr6_-_39290744 | 0.63 |
ENST00000507712.1
|
KCNK16
|
potassium channel, subfamily K, member 16 |
| chrX_-_64196376 | 0.62 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chrX_-_64196307 | 0.58 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr11_+_5410607 | 0.58 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr3_+_111717600 | 0.58 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr7_+_73507409 | 0.58 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr3_+_111717511 | 0.57 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chrX_-_64196351 | 0.56 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr8_-_145018905 | 0.55 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr3_+_47021168 | 0.55 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr20_-_48747662 | 0.53 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr14_-_106586656 | 0.53 |
ENST00000390602.2
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr16_+_23194033 | 0.52 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr17_+_61086917 | 0.52 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_+_129847688 | 0.51 |
ENST00000297819.3
|
SSMEM1
|
serine-rich single-pass membrane protein 1 |
| chr15_-_20170354 | 0.51 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr7_+_30634297 | 0.50 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr19_+_34856141 | 0.50 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr2_-_136288740 | 0.50 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr4_+_71200681 | 0.50 |
ENST00000273936.5
|
CABS1
|
calcium-binding protein, spermatid-specific 1 |
| chr4_+_37245799 | 0.50 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr6_+_41010293 | 0.49 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr13_-_46756351 | 0.49 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_+_116782527 | 0.49 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr16_-_20681177 | 0.49 |
ENST00000524149.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr10_+_74653330 | 0.48 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr4_-_123377880 | 0.47 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chrX_+_41548220 | 0.47 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr14_-_107049312 | 0.47 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr3_-_58613323 | 0.47 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr1_+_228337553 | 0.47 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr4_-_69111401 | 0.46 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chrX_+_41548259 | 0.46 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr1_+_16348525 | 0.45 |
ENST00000331433.4
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr12_+_66218903 | 0.44 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr16_+_58426296 | 0.44 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr7_-_766879 | 0.44 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_-_99057551 | 0.43 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr2_-_46385 | 0.43 |
ENST00000327669.4
|
FAM110C
|
family with sequence similarity 110, member C |
| chr2_-_110371720 | 0.43 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr6_-_32145861 | 0.42 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr11_+_64808675 | 0.42 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr12_+_9144626 | 0.41 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_+_158223923 | 0.41 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr5_+_14664762 | 0.39 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr16_+_32063311 | 0.39 |
ENST00000426099.1
|
AC142381.1
|
AC142381.1 |
| chr16_+_32077386 | 0.39 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr3_-_116163830 | 0.38 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr14_+_24540046 | 0.38 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr14_+_22356029 | 0.38 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr9_-_21166659 | 0.37 |
ENST00000380225.1
|
IFNA21
|
interferon, alpha 21 |
| chr14_-_106068065 | 0.37 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr17_-_33390667 | 0.37 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr4_-_153601136 | 0.36 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr12_+_121416489 | 0.35 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr7_+_112063192 | 0.35 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr14_-_106552755 | 0.35 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr3_-_178984759 | 0.35 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr2_-_106054952 | 0.33 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr8_+_67405755 | 0.33 |
ENST00000521495.1
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr1_-_155006224 | 0.33 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr19_-_18391708 | 0.32 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr12_-_102591604 | 0.32 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr22_-_32341336 | 0.32 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr7_-_99277610 | 0.32 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr2_-_68547061 | 0.32 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr19_+_2249308 | 0.31 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr1_-_95007193 | 0.31 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr14_-_106692191 | 0.30 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_+_12040238 | 0.30 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr9_-_22009297 | 0.30 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr1_+_173793777 | 0.30 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr4_-_170947565 | 0.30 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr11_-_72353451 | 0.30 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr5_-_101834617 | 0.30 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr10_+_71078595 | 0.29 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr6_+_34433844 | 0.29 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr4_+_144303093 | 0.29 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_-_101834712 | 0.28 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr12_-_11548496 | 0.28 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr14_-_58894332 | 0.28 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr16_+_33006369 | 0.27 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr16_+_33629600 | 0.27 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr14_-_107131560 | 0.27 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr4_-_819901 | 0.27 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr12_-_75603643 | 0.26 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr12_+_66218598 | 0.26 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr17_+_71228793 | 0.25 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr5_-_176056974 | 0.25 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr12_-_75603482 | 0.25 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr12_-_75603202 | 0.25 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr9_-_21202204 | 0.25 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr3_-_53164423 | 0.25 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr2_-_29297127 | 0.25 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr1_-_67266939 | 0.24 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr22_+_38609538 | 0.24 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chrX_-_53461288 | 0.24 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_+_74011328 | 0.23 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr6_-_32806506 | 0.23 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr14_-_58894223 | 0.23 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr5_+_149737202 | 0.23 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr2_-_132249955 | 0.23 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr13_-_103346854 | 0.22 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr16_+_89696692 | 0.22 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr12_+_7941989 | 0.22 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr7_-_36764142 | 0.22 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr9_-_128412696 | 0.22 |
ENST00000420643.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr14_+_22670455 | 0.22 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr14_+_92980111 | 0.21 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr1_+_151512775 | 0.21 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr3_-_16524357 | 0.20 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr12_-_75603236 | 0.20 |
ENST00000540018.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_+_207024306 | 0.20 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chrX_+_72064690 | 0.19 |
ENST00000438696.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr19_-_11347173 | 0.19 |
ENST00000587656.1
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr3_-_73673991 | 0.19 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr14_-_107199464 | 0.19 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr2_-_55277692 | 0.18 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr12_+_66218212 | 0.18 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr2_-_55277654 | 0.18 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr3_+_52812523 | 0.18 |
ENST00000540715.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr2_-_207024134 | 0.18 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_+_52813932 | 0.17 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr14_-_106963409 | 0.17 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr1_+_224301787 | 0.17 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr22_-_37213554 | 0.17 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr3_-_196242233 | 0.17 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr22_+_38453207 | 0.17 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr18_-_73139589 | 0.16 |
ENST00000382638.3
ENST00000584508.1 ENST00000579022.1 |
SMIM21
|
small integral membrane protein 21 |
| chr12_+_60058458 | 0.15 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr6_+_125524785 | 0.15 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr10_-_15762124 | 0.15 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr20_+_42143136 | 0.15 |
ENST00000373134.1
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr6_+_111408698 | 0.15 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr8_+_128426535 | 0.15 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr13_+_46039037 | 0.14 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr16_+_29674540 | 0.14 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr14_-_107219365 | 0.14 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr4_+_141264597 | 0.13 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr17_+_39994032 | 0.13 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr1_+_74701062 | 0.13 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr12_-_5352315 | 0.13 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr2_-_207023918 | 0.13 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr8_+_24241789 | 0.13 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr5_+_3596168 | 0.12 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr12_-_21810765 | 0.12 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr3_+_63953415 | 0.12 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr9_-_21239978 | 0.12 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr16_-_33647696 | 0.11 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr12_-_95397442 | 0.11 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr11_-_19223523 | 0.11 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr18_-_13915530 | 0.11 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr21_-_35899113 | 0.11 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr12_-_21810726 | 0.11 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr16_-_75301886 | 0.11 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr3_-_139195350 | 0.11 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr2_-_55277436 | 0.10 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr6_-_133084580 | 0.10 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr4_+_15341442 | 0.10 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr5_-_78281775 | 0.10 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr5_+_80529104 | 0.10 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr3_+_67048721 | 0.09 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr10_-_118032697 | 0.09 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.6 | 2.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.5 | 1.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.4 | 1.5 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 0.9 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.3 | 0.3 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) |
| 0.3 | 0.9 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 0.8 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 0.8 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.9 | GO:2000685 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.2 | 0.8 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.5 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.2 | 3.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.5 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 3.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.7 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 3.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 3.3 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 1.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 2.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 5.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 3.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 1.4 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.3 | 3.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.9 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.2 | 0.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.2 | 0.8 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.5 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 2.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 3.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |