Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
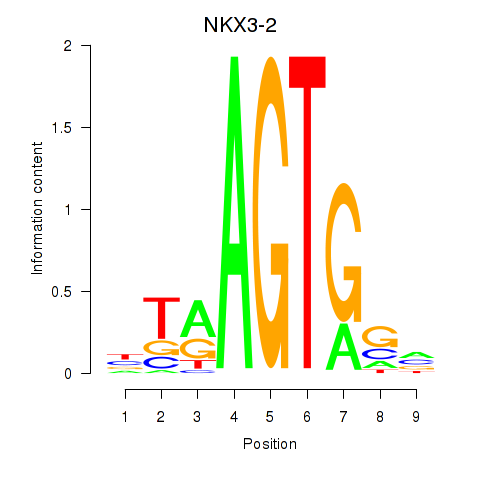
Results for NKX3-2
Z-value: 1.29
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | -0.21 | 2.6e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_112970436 | 12.16 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr1_-_161337662 | 11.59 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr9_-_34397800 | 10.71 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr5_-_149792295 | 7.94 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr6_-_32557610 | 7.62 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr7_-_99573677 | 7.37 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr17_-_74137374 | 7.26 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr13_+_43355683 | 6.96 |
ENST00000537894.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr13_+_43355732 | 6.94 |
ENST00000313851.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr9_+_34458771 | 6.94 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr11_-_111175739 | 5.61 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr5_+_156696362 | 4.72 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_183622442 | 4.70 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr6_+_32821924 | 4.53 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr16_-_21289627 | 4.52 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr2_+_98703643 | 4.40 |
ENST00000477737.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr16_+_57406368 | 4.24 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr17_+_68071389 | 4.21 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_-_26593649 | 4.21 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr19_+_41497178 | 4.10 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr11_-_26593779 | 3.90 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr9_-_75567962 | 3.83 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr7_-_99573640 | 3.73 |
ENST00000411734.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_+_162351503 | 3.72 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr4_-_168155730 | 3.68 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_125694347 | 3.61 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr4_-_141348789 | 3.49 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr4_-_141348999 | 3.43 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr9_-_124976185 | 3.41 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr9_-_124976154 | 3.34 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr5_+_156712372 | 3.24 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_-_112693865 | 3.09 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr11_-_26593677 | 3.03 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_113658892 | 2.94 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr19_-_6720686 | 2.86 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr6_-_32731299 | 2.77 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr7_-_120498357 | 2.71 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr15_-_40401062 | 2.71 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr4_-_168155417 | 2.70 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_-_54468974 | 2.63 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr3_-_112693759 | 2.62 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr1_-_217262933 | 2.62 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr4_-_168155700 | 2.60 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_7745232 | 2.56 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_+_68071458 | 2.55 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_217262969 | 2.49 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr5_-_160279207 | 2.47 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr12_+_56075330 | 2.38 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr3_+_169629354 | 2.36 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr15_-_43513187 | 2.23 |
ENST00000540029.1
ENST00000441366.2 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr17_+_58755184 | 2.22 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr7_+_90338712 | 2.22 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr6_+_159084188 | 2.19 |
ENST00000367081.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr1_-_216978709 | 2.12 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr10_-_46168156 | 2.08 |
ENST00000374371.2
ENST00000335258.7 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr2_-_220435963 | 2.08 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr19_-_3028354 | 2.03 |
ENST00000586422.1
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr5_-_176326333 | 1.92 |
ENST00000292432.5
|
HK3
|
hexokinase 3 (white cell) |
| chr11_-_106889157 | 1.87 |
ENST00000282249.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr4_-_168155577 | 1.75 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_+_89402148 | 1.74 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr11_+_66045634 | 1.74 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr1_+_211433275 | 1.70 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr14_-_23770683 | 1.70 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr6_+_18155560 | 1.70 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr11_-_59633951 | 1.67 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr16_-_3350614 | 1.65 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr3_-_112218378 | 1.63 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr10_+_35415851 | 1.62 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr4_+_17579110 | 1.60 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chrX_+_48660287 | 1.59 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr1_-_217250231 | 1.57 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr3_+_132316081 | 1.56 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr19_+_17337473 | 1.54 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr2_-_220436248 | 1.53 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr2_-_25194963 | 1.53 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr12_-_10151773 | 1.52 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr1_+_60280458 | 1.52 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr7_+_90339169 | 1.50 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr19_-_51412584 | 1.48 |
ENST00000431178.2
|
KLK4
|
kallikrein-related peptidase 4 |
| chr12_-_42983478 | 1.46 |
ENST00000345127.3
ENST00000547113.1 |
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr1_-_48866517 | 1.40 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chrX_+_95939711 | 1.40 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr11_+_36317830 | 1.33 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr12_+_56473939 | 1.32 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_+_18380051 | 1.30 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr6_+_29910301 | 1.30 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr11_-_63381823 | 1.29 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr14_+_21156915 | 1.27 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr3_-_112218205 | 1.26 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr2_-_188312971 | 1.25 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr8_+_67624653 | 1.24 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr4_+_159727272 | 1.23 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_+_3019552 | 1.23 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr3_-_160117301 | 1.22 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr14_+_104604916 | 1.21 |
ENST00000423312.2
|
KIF26A
|
kinesin family member 26A |
| chr1_-_156399184 | 1.19 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr11_+_60970852 | 1.19 |
ENST00000325558.6
|
PGA3
|
pepsinogen 3, group I (pepsinogen A) |
| chr11_+_118978045 | 1.19 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr2_-_208489707 | 1.18 |
ENST00000448007.2
ENST00000432416.1 ENST00000411432.1 |
METTL21A
|
methyltransferase like 21A |
| chr7_-_50628745 | 1.17 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr6_-_116381918 | 1.16 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr21_-_43816052 | 1.14 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr2_+_186603545 | 1.13 |
ENST00000424728.1
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr11_-_22851367 | 1.13 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr6_+_29274403 | 1.10 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr6_-_27279949 | 1.07 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr11_-_77531752 | 1.04 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr20_+_53092123 | 1.03 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr5_-_36152031 | 1.02 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr17_-_9929581 | 1.02 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr6_-_31697255 | 1.01 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr10_+_16478942 | 1.01 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr20_-_2821271 | 1.00 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr15_+_90895471 | 0.99 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr19_-_7764281 | 0.98 |
ENST00000360067.4
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr12_+_6309517 | 0.96 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr8_+_144295067 | 0.96 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr1_+_160097462 | 0.96 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr3_+_48507210 | 0.95 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr14_+_50779071 | 0.94 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr10_-_27529486 | 0.94 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr11_-_102401469 | 0.94 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr3_-_52443799 | 0.94 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr1_+_158259558 | 0.94 |
ENST00000368170.3
|
CD1C
|
CD1c molecule |
| chr19_+_54609230 | 0.94 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr12_-_122712038 | 0.94 |
ENST00000413918.1
ENST00000443649.3 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr14_-_68066849 | 0.92 |
ENST00000558493.1
ENST00000561272.1 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr2_-_25016251 | 0.92 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr5_+_56111361 | 0.92 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr20_+_53092232 | 0.92 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chrX_+_76709648 | 0.92 |
ENST00000439435.1
|
FGF16
|
fibroblast growth factor 16 |
| chr20_-_2821756 | 0.91 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr14_-_92506371 | 0.91 |
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr15_+_88120158 | 0.88 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr20_+_2276639 | 0.88 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr1_-_202858227 | 0.87 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr10_-_127505167 | 0.85 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr7_-_56160666 | 0.84 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr12_+_56473910 | 0.84 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_-_165324983 | 0.84 |
ENST00000367893.4
|
LMX1A
|
LIM homeobox transcription factor 1, alpha |
| chr16_-_86532148 | 0.84 |
ENST00000594398.1
|
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr14_-_68000442 | 0.83 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr20_+_15177480 | 0.81 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr3_+_52454971 | 0.81 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr20_+_55966444 | 0.81 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr1_+_210001309 | 0.79 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr20_-_34330129 | 0.79 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr4_-_11430221 | 0.78 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr1_+_22307592 | 0.77 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr5_+_64859066 | 0.77 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr10_+_35416223 | 0.77 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr22_-_21356375 | 0.77 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr10_-_27529716 | 0.75 |
ENST00000375897.3
ENST00000396271.3 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chrX_-_15332665 | 0.75 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr11_-_116662593 | 0.75 |
ENST00000227665.4
|
APOA5
|
apolipoprotein A-V |
| chr1_-_91487013 | 0.74 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr20_-_61557821 | 0.74 |
ENST00000354665.4
ENST00000370368.1 ENST00000395343.1 ENST00000395340.1 |
DIDO1
|
death inducer-obliterator 1 |
| chr3_-_146262293 | 0.74 |
ENST00000448205.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_+_6845497 | 0.73 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr16_+_5121814 | 0.73 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr15_-_43559055 | 0.71 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr17_-_7145475 | 0.71 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr19_+_16999654 | 0.71 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr3_-_52008016 | 0.70 |
ENST00000489595.2
ENST00000461108.1 ENST00000395008.2 ENST00000525795.1 ENST00000361143.5 |
RP11-155D18.14
ABHD14B
|
Poly(rC)-binding protein 4 abhydrolase domain containing 14B |
| chr21_-_33984888 | 0.70 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr2_-_24270217 | 0.70 |
ENST00000295148.4
ENST00000406895.3 |
C2orf44
|
chromosome 2 open reading frame 44 |
| chr17_-_79623597 | 0.70 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr1_+_35734562 | 0.69 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr7_-_56160625 | 0.69 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr7_-_79082867 | 0.68 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_+_23847432 | 0.68 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr15_-_37393406 | 0.68 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_-_77531858 | 0.68 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr9_+_134065506 | 0.67 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr9_+_35161998 | 0.66 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chr8_-_27115931 | 0.66 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr17_-_42295870 | 0.66 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr5_+_68710906 | 0.65 |
ENST00000325631.5
ENST00000454295.2 |
MARVELD2
|
MARVEL domain containing 2 |
| chr21_-_33984865 | 0.64 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr10_-_49459800 | 0.64 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr9_+_5890802 | 0.63 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chrX_-_71526813 | 0.63 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr2_+_232572361 | 0.62 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr18_-_6414884 | 0.61 |
ENST00000317931.7
ENST00000284898.6 ENST00000400104.3 |
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr19_-_10444188 | 0.61 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr17_-_60883993 | 0.60 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_-_3061397 | 0.60 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr5_+_169780485 | 0.60 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr11_+_77532155 | 0.60 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr19_-_36545649 | 0.59 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr6_-_30080876 | 0.59 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr1_+_156756667 | 0.59 |
ENST00000526188.1
ENST00000454659.1 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr1_-_154164534 | 0.58 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr6_-_47010061 | 0.57 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr19_+_18544045 | 0.56 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_-_77532050 | 0.56 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr5_+_68711209 | 0.56 |
ENST00000512803.1
|
MARVELD2
|
MARVEL domain containing 2 |
| chr9_-_32526184 | 0.56 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr7_+_73106926 | 0.55 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.6 | 7.9 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.5 | 7.6 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.4 | 4.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.0 | 3.8 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.0 | 2.9 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.8 | 8.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.6 | 1.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.5 | 6.8 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 | 10.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 1.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.4 | 1.6 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.4 | 1.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.4 | 1.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 1.5 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.3 | 6.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 4.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.5 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 0.9 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.3 | 0.9 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.3 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 2.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 1.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.9 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 9.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.2 | 4.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 1.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 2.9 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.2 | 2.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 2.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 1.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 1.0 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.2 | 1.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 2.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 1.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 11.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 0.7 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.5 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 3.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 7.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 6.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.7 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.4 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.7 | GO:2000302 | regulation of synaptic vesicle priming(GO:0010807) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.9 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 10.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.5 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 1.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 2.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 2.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 2.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 1.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 1.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 2.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 2.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.9 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.3 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.5 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.7 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.9 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.2 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 7.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 1.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 1.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.5 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 2.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 4.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 2.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 1.4 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 1.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.8 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 1.7 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0060701 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.0 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.7 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.6 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) positive regulation of estrogen secretion(GO:2000863) regulation of estradiol secretion(GO:2000864) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0008037 | cell recognition(GO:0008037) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.7 | 3.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 6.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.6 | 2.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.5 | 3.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 4.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 10.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 1.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 0.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 1.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.2 | 1.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.5 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 13.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 2.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 4.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 8.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 3.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 4.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 9.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.3 | 8.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.1 | 4.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.8 | 3.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.7 | 2.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 11.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.4 | 1.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 10.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 6.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.3 | 2.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 6.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 3.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 2.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 1.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 1.9 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 7.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 4.1 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 2.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 3.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 6.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 2.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 1.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 4.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.7 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 3.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 2.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 3.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 3.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 3.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 4.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 1.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.1 | 0.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 5.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 5.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 7.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 3.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 6.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 7.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 8.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 5.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |