Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
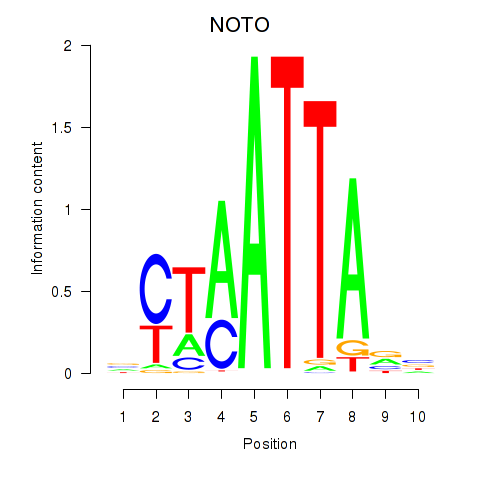
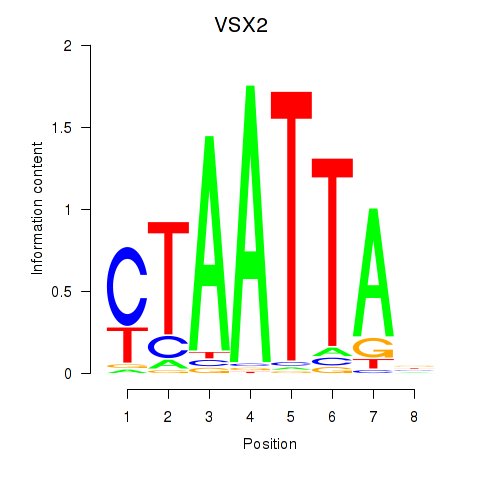
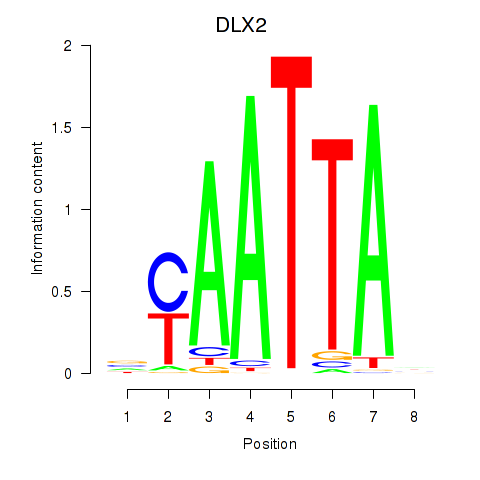
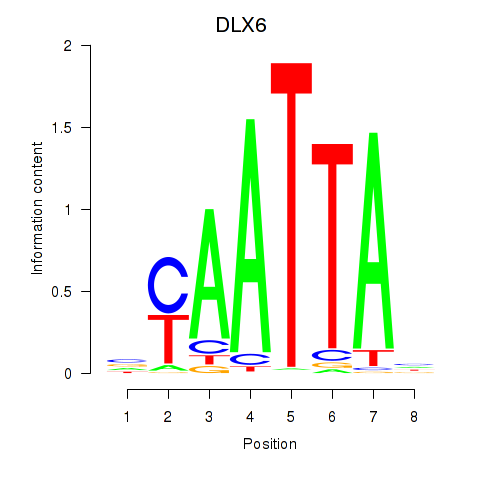
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.64
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NOTO
|
ENSG00000214513.3 | notochord homeobox |
|
VSX2
|
ENSG00000119614.2 | visual system homeobox 2 |
|
DLX2
|
ENSG00000115844.6 | distal-less homeobox 2 |
|
DLX6
|
ENSG00000006377.9 | distal-less homeobox 6 |
|
NKX6-1
|
ENSG00000163623.5 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX2 | hg19_v2_chr2_-_172967621_172967637 | 0.54 | 2.2e-03 | Click! |
| DLX6 | hg19_v2_chr7_+_96634850_96634874, hg19_v2_chr7_+_96635277_96635301 | -0.29 | 1.2e-01 | Click! |
| NKX6-1 | hg19_v2_chr4_-_85418103_85418103 | 0.15 | 4.3e-01 | Click! |
Activity profile of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Sorted Z-values of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_36050881 | 4.03 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr17_-_39203519 | 3.46 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr6_-_87804815 | 2.99 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr7_-_25268104 | 2.49 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr1_-_183622442 | 2.09 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr8_-_10512569 | 1.92 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr20_-_7921090 | 1.61 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr4_-_138453606 | 1.57 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr2_-_68052694 | 1.48 |
ENST00000457448.1
|
AC010987.6
|
AC010987.6 |
| chr6_-_76203345 | 1.47 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr6_-_76203454 | 1.42 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr20_-_50419055 | 1.34 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_-_31531121 | 1.33 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr4_+_71108300 | 1.28 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr1_+_183774240 | 1.23 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr16_-_28621353 | 1.21 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_-_145278475 | 1.20 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr14_-_92413727 | 1.18 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr1_-_150738261 | 1.15 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr12_-_16761007 | 1.14 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr16_+_53133070 | 1.14 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_-_54423529 | 1.09 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr4_-_25865159 | 1.06 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr9_-_215744 | 1.03 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr8_-_72268968 | 1.00 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr14_-_92413353 | 0.99 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chrX_-_73512411 | 0.97 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr8_-_72268889 | 0.97 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrX_-_44202857 | 0.94 |
ENST00000420999.1
|
EFHC2
|
EF-hand domain (C-terminal) containing 2 |
| chr1_+_244515930 | 0.92 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr9_+_12693336 | 0.89 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr5_-_137475071 | 0.82 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr3_+_111717600 | 0.78 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr1_+_61548374 | 0.78 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr11_-_40315640 | 0.78 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr3_+_111718036 | 0.77 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr17_-_10421853 | 0.77 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr18_+_32173276 | 0.76 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr15_-_37393406 | 0.76 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr12_+_54378923 | 0.76 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr7_-_130080977 | 0.75 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr4_+_77356248 | 0.74 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr12_-_68696652 | 0.73 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr12_+_26348582 | 0.73 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr20_-_50418947 | 0.73 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr5_-_24645078 | 0.73 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr3_+_111718173 | 0.72 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr5_-_96478466 | 0.71 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr3_+_111717511 | 0.70 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr5_+_156696362 | 0.68 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_7698599 | 0.68 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr3_+_2933893 | 0.67 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr19_-_56110859 | 0.66 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr6_-_161695042 | 0.65 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr20_-_50418972 | 0.64 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_21494654 | 0.62 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr17_-_39165366 | 0.61 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr8_-_133637624 | 0.61 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr13_-_70682590 | 0.59 |
ENST00000377844.4
|
KLHL1
|
kelch-like family member 1 |
| chr4_-_177116772 | 0.58 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr17_-_39280419 | 0.58 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr6_+_135502501 | 0.58 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_-_33913708 | 0.56 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr17_+_68071389 | 0.54 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_+_53751445 | 0.54 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr2_+_128403439 | 0.54 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr7_+_99717230 | 0.54 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr11_-_129062093 | 0.53 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr5_-_13944652 | 0.53 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr6_-_161695074 | 0.52 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_+_128403720 | 0.52 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr17_-_77924627 | 0.52 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_+_61548225 | 0.52 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr3_-_114477787 | 0.52 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_76072719 | 0.51 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr6_-_22297730 | 0.50 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr3_+_69928256 | 0.50 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr8_-_109799793 | 0.49 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr8_+_50824233 | 0.49 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr10_-_102989551 | 0.49 |
ENST00000370193.2
|
LBX1
|
ladybird homeobox 1 |
| chr7_+_23286182 | 0.49 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr18_-_53089723 | 0.49 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr3_-_141747950 | 0.48 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_94586651 | 0.48 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr1_+_180601139 | 0.48 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr13_-_84456527 | 0.47 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr11_+_71900703 | 0.47 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chrX_-_46187069 | 0.47 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr4_-_123542224 | 0.47 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr11_+_71900572 | 0.46 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr12_-_52946923 | 0.46 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr6_+_39760129 | 0.46 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr3_-_114477962 | 0.45 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_-_36050819 | 0.45 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr7_+_73442487 | 0.45 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr11_+_124481361 | 0.44 |
ENST00000284288.2
|
PANX3
|
pannexin 3 |
| chr18_-_47792851 | 0.44 |
ENST00000398545.4
|
CCDC11
|
coiled-coil domain containing 11 |
| chrX_-_117119243 | 0.44 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr4_-_138453559 | 0.44 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr7_+_116660246 | 0.44 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr2_-_203735586 | 0.43 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr4_-_74486109 | 0.43 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_-_124190184 | 0.43 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr13_-_46716969 | 0.42 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_-_16759711 | 0.42 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr19_-_7697857 | 0.41 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr1_+_61547405 | 0.40 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr21_+_17442799 | 0.40 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_41884582 | 0.39 |
ENST00000499082.2
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr11_-_26593779 | 0.39 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_+_71494461 | 0.39 |
ENST00000396073.3
|
ENAM
|
enamelin |
| chr13_-_41593425 | 0.38 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_61547894 | 0.38 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr21_-_32253874 | 0.38 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr3_+_173116225 | 0.38 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_+_41614909 | 0.37 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chrX_-_10851762 | 0.36 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr4_+_71458012 | 0.36 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr7_+_73442422 | 0.35 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr2_+_162272605 | 0.35 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr3_+_113616317 | 0.35 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr15_+_76352178 | 0.34 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr19_+_50016411 | 0.34 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_-_40337470 | 0.34 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr12_+_41831485 | 0.33 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr4_+_66536248 | 0.33 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr1_-_48937838 | 0.33 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr9_+_2159850 | 0.32 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_121986923 | 0.32 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr12_-_86650045 | 0.32 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_-_76782371 | 0.32 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr2_+_216946589 | 0.32 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr9_+_130026756 | 0.31 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr4_+_159727272 | 0.31 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr12_+_20963647 | 0.31 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_-_111804905 | 0.31 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_+_72783026 | 0.31 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr21_-_31538971 | 0.30 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr17_+_59489112 | 0.30 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr1_+_101003687 | 0.30 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr2_-_216946500 | 0.30 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr20_+_11008408 | 0.30 |
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187 |
| chr20_+_52105495 | 0.30 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr6_-_9933500 | 0.30 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr15_+_41913690 | 0.30 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr11_-_102496063 | 0.29 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr12_+_20963632 | 0.29 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr13_+_108922228 | 0.29 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr5_+_69321074 | 0.29 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr16_-_28621312 | 0.29 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr1_-_227505289 | 0.29 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr1_+_44679113 | 0.29 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr3_+_120626919 | 0.29 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr4_-_105416039 | 0.28 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr17_+_39261584 | 0.28 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chrX_+_68835911 | 0.28 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr14_-_69261310 | 0.28 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_-_26593649 | 0.28 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr7_+_141478242 | 0.27 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr6_-_33160231 | 0.27 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr8_-_72274095 | 0.27 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr8_+_121137333 | 0.27 |
ENST00000309791.4
ENST00000297848.3 ENST00000247781.3 |
COL14A1
|
collagen, type XIV, alpha 1 |
| chr17_-_72527605 | 0.27 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr4_-_74486347 | 0.27 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr21_+_17443434 | 0.27 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_40713573 | 0.27 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr19_+_50016610 | 0.26 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_114790179 | 0.26 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr5_-_160279207 | 0.26 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr11_+_20620946 | 0.26 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr6_+_5261225 | 0.26 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr10_-_62332357 | 0.26 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_+_107110488 | 0.25 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr2_+_179184955 | 0.25 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr10_+_111765562 | 0.25 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr2_+_234959376 | 0.25 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr6_-_32908792 | 0.25 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr9_-_136933134 | 0.25 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr10_-_1779663 | 0.25 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr5_-_35195338 | 0.25 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chr7_+_73442102 | 0.24 |
ENST00000445912.1
ENST00000252034.7 |
ELN
|
elastin |
| chr3_+_157154578 | 0.24 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr18_-_53069419 | 0.24 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr7_-_92855762 | 0.24 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr11_+_15136462 | 0.24 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr14_-_98444461 | 0.24 |
ENST00000499006.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr13_-_36788718 | 0.23 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr9_-_124989804 | 0.23 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr13_-_88323218 | 0.23 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr7_+_114055052 | 0.23 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr3_-_72149553 | 0.23 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr11_-_26593677 | 0.23 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr5_+_133562095 | 0.23 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr3_-_27763803 | 0.23 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr6_+_12717892 | 0.22 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr7_+_97361388 | 0.22 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr2_+_183943464 | 0.22 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr3_-_112360116 | 0.22 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr4_+_89206076 | 0.22 |
ENST00000500009.2
|
RP11-10L7.1
|
RP11-10L7.1 |
| chr5_-_159827033 | 0.22 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NOTO_VSX2_DLX2_DLX6_NKX6-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048392 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 0.9 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.3 | 2.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 2.6 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.3 | 1.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 | 0.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 0.6 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.2 | 2.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 2.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.9 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 1.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.5 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.3 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 2.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 1.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.2 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.8 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.6 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0003150 | membranous septum morphogenesis(GO:0003149) muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 3.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 2.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 1.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.8 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 2.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |