Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
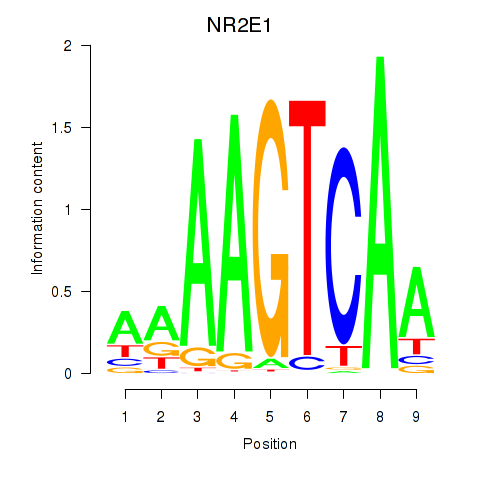
Results for NR2E1
Z-value: 0.81
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | 0.25 | 1.8e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91539918 | 2.83 |
ENST00000548218.1
|
DCN
|
decorin |
| chr2_-_31637560 | 2.19 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_+_113885138 | 1.90 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr5_-_11588907 | 1.85 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr1_-_205419053 | 1.83 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr4_-_47983519 | 1.65 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr1_+_32042105 | 1.58 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr11_-_62323702 | 1.51 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr18_+_29027696 | 1.48 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr4_-_111119804 | 1.43 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr5_-_11589131 | 1.42 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr11_-_63439381 | 1.35 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr12_-_82152444 | 1.24 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr10_+_11784360 | 1.22 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr17_-_41623691 | 1.14 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr12_-_91572278 | 1.01 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr11_-_63439013 | 1.00 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chrX_-_65253506 | 0.97 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr11_+_35198243 | 0.97 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_+_116166331 | 0.97 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr6_+_74405501 | 0.96 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr3_+_121774202 | 0.95 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr7_+_80267973 | 0.94 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_+_56915776 | 0.94 |
ENST00000550726.1
ENST00000542360.1 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr7_-_107443652 | 0.93 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr1_-_162381907 | 0.90 |
ENST00000367929.2
ENST00000359567.3 |
SH2D1B
|
SH2 domain containing 1B |
| chr6_+_12290586 | 0.89 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr22_-_30695471 | 0.88 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chr17_-_41623716 | 0.88 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr11_-_111794446 | 0.87 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr14_+_22977587 | 0.86 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chrX_+_47444613 | 0.86 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr16_-_20709066 | 0.82 |
ENST00000520010.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr6_-_66417107 | 0.82 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr2_-_85637459 | 0.82 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr9_-_21994344 | 0.80 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr7_-_121944491 | 0.80 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr8_-_16859690 | 0.80 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chrX_-_65259900 | 0.80 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr12_+_56915713 | 0.79 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr2_+_90077680 | 0.78 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr3_+_136676707 | 0.78 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr2_-_89442621 | 0.78 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr17_+_56232494 | 0.78 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr4_-_4543700 | 0.77 |
ENST00000505286.1
ENST00000306200.2 |
STX18
|
syntaxin 18 |
| chr4_+_71063641 | 0.77 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chrX_+_135614293 | 0.77 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chrX_-_65259914 | 0.76 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr9_-_110251836 | 0.76 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr3_-_116163830 | 0.75 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_-_63439174 | 0.74 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr2_+_101437487 | 0.72 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_-_208030647 | 0.72 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_28124903 | 0.71 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_+_124320195 | 0.71 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_-_158300556 | 0.70 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr14_+_67291158 | 0.69 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr1_+_25944341 | 0.68 |
ENST00000263979.3
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr10_+_124320156 | 0.67 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr6_-_134861089 | 0.66 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr2_+_135011731 | 0.66 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr16_-_15180257 | 0.65 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr17_-_76220740 | 0.65 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr19_+_11546153 | 0.64 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr2_+_11679963 | 0.63 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr12_+_53662073 | 0.62 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr12_+_53662110 | 0.62 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr9_-_99064386 | 0.62 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr17_+_17082842 | 0.62 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr19_+_11546093 | 0.61 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr6_-_112080256 | 0.61 |
ENST00000462856.2
ENST00000229471.4 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr1_+_214163033 | 0.59 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr12_-_95044309 | 0.59 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr12_-_52887034 | 0.58 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr3_+_189349162 | 0.56 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr6_+_106546808 | 0.56 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_+_52812523 | 0.54 |
ENST00000540715.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr17_-_40833858 | 0.53 |
ENST00000332438.4
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chrX_-_53711064 | 0.52 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr5_-_179047881 | 0.52 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr19_+_11546440 | 0.52 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr6_+_125540951 | 0.49 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr17_-_46667594 | 0.49 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr3_+_156799587 | 0.49 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr6_-_88411911 | 0.48 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr12_+_14518598 | 0.48 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_192141611 | 0.47 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr3_+_141106458 | 0.47 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_11666649 | 0.46 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr17_+_1959369 | 0.46 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr12_-_82152420 | 0.46 |
ENST00000552948.1
ENST00000548586.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_-_54824673 | 0.45 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr6_+_15246501 | 0.44 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chrX_-_49041242 | 0.44 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr11_-_66104237 | 0.44 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr4_+_96012614 | 0.44 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_+_47078069 | 0.44 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr11_-_128457446 | 0.43 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr1_+_40420802 | 0.43 |
ENST00000372811.5
ENST00000420632.2 ENST00000434861.1 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr1_+_77333117 | 0.43 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_-_157892055 | 0.43 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr6_-_130543958 | 0.43 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr10_-_29084886 | 0.42 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr1_+_165864800 | 0.42 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr10_+_52152766 | 0.42 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr11_-_111781454 | 0.42 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_140013275 | 0.41 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr9_+_99691286 | 0.41 |
ENST00000372322.3
|
NUTM2G
|
NUT family member 2G |
| chr5_-_127873659 | 0.41 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chrX_+_115301975 | 0.40 |
ENST00000371906.4
|
AGTR2
|
angiotensin II receptor, type 2 |
| chr3_+_42897512 | 0.40 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr10_-_21786179 | 0.40 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr10_-_21463116 | 0.40 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr5_-_82969405 | 0.40 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_209010874 | 0.39 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr3_+_52813932 | 0.38 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr16_+_3068393 | 0.38 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr1_-_154155595 | 0.38 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr18_+_22040620 | 0.38 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr17_+_12569306 | 0.38 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr12_-_10324716 | 0.37 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_-_124408652 | 0.37 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr1_-_197036364 | 0.37 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr9_-_97090926 | 0.37 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr1_+_154975258 | 0.36 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr15_+_63188009 | 0.36 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_+_115572415 | 0.36 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr1_-_20834586 | 0.36 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr17_-_46667628 | 0.35 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr14_-_57277163 | 0.35 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr17_-_47925379 | 0.35 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr4_-_52883786 | 0.35 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr11_-_111781554 | 0.34 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr12_-_8088871 | 0.34 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr17_-_73975444 | 0.34 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr1_-_31661000 | 0.33 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chrX_+_47077632 | 0.33 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr9_-_99064429 | 0.33 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr12_-_99548645 | 0.33 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr8_+_11561660 | 0.33 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr6_-_9977801 | 0.33 |
ENST00000316020.6
ENST00000491508.1 |
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr3_-_64673668 | 0.33 |
ENST00000498707.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr12_-_91546926 | 0.33 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_-_13546632 | 0.33 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chrX_-_124097620 | 0.33 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr19_-_47220335 | 0.32 |
ENST00000601806.1
ENST00000593363.1 ENST00000598633.1 ENST00000595515.1 ENST00000433867.1 |
PRKD2
|
protein kinase D2 |
| chr18_+_22040593 | 0.32 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr17_-_41738931 | 0.32 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr14_+_24407940 | 0.32 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr1_-_154150651 | 0.32 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chrX_-_33146477 | 0.32 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr19_-_47157914 | 0.31 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_14026063 | 0.31 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr17_+_35732916 | 0.31 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr2_-_106013400 | 0.31 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr4_-_153303658 | 0.31 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_-_154155675 | 0.31 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr3_-_54962100 | 0.31 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr13_-_21634421 | 0.31 |
ENST00000542899.1
|
LATS2
|
large tumor suppressor kinase 2 |
| chr8_+_77593448 | 0.30 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_-_30310797 | 0.30 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr1_+_248201474 | 0.30 |
ENST00000366479.2
|
OR2L2
|
olfactory receptor, family 2, subfamily L, member 2 |
| chr17_+_35732955 | 0.30 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr5_+_9546306 | 0.29 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr10_-_115423792 | 0.29 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr2_-_183387064 | 0.29 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_+_161691353 | 0.28 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr5_-_135290705 | 0.28 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_1781906 | 0.28 |
ENST00000453348.1
ENST00000415399.1 |
AC074389.9
|
AC074389.9 |
| chr1_+_225600404 | 0.28 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr5_-_127873496 | 0.28 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr5_+_32711829 | 0.28 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr17_+_29664830 | 0.27 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr14_-_57277178 | 0.27 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr3_+_12392971 | 0.27 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr8_-_93029520 | 0.26 |
ENST00000521553.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_+_110656005 | 0.26 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr6_+_31515337 | 0.26 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr1_-_185597619 | 0.26 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr20_+_62795827 | 0.26 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr14_+_51955831 | 0.26 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr3_-_33686925 | 0.25 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_+_22636283 | 0.25 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chr17_-_66951474 | 0.25 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr7_-_16505440 | 0.25 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr1_-_216596738 | 0.25 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr1_-_166944561 | 0.25 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr9_-_95244781 | 0.25 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr6_-_155776966 | 0.25 |
ENST00000159060.2
|
NOX3
|
NADPH oxidase 3 |
| chr6_+_107077435 | 0.24 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr2_-_214014959 | 0.24 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr12_-_99288536 | 0.24 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_81187753 | 0.24 |
ENST00000312465.7
ENST00000456523.3 |
FGF5
|
fibroblast growth factor 5 |
| chr14_+_69865401 | 0.23 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr3_-_64673289 | 0.23 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr8_-_124037890 | 0.23 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr20_+_42574317 | 0.23 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr16_-_69788816 | 0.23 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chrX_-_105282712 | 0.22 |
ENST00000372563.1
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_-_107880508 | 0.22 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.3 | 1.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 1.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 1.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 | 0.9 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 4.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.8 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 0.8 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.2 | 0.7 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 1.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.8 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 0.8 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 1.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.4 | GO:0071726 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 0.6 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.8 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.3 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0021546 | rhombomere development(GO:0021546) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.4 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 2.5 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 0.8 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.7 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.0 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.3 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.3 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.1 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 3.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.6 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 2.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0048690 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.7 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0070257 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 0.8 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.2 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 3.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 2.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.3 | 1.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 2.9 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.7 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 1.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.7 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 5.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |