Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for NR4A1
Z-value: 0.58
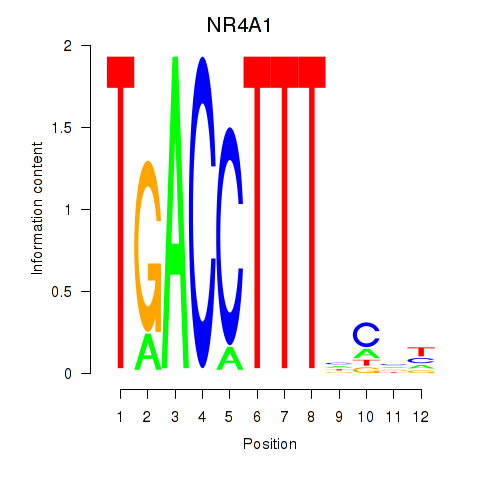
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | nuclear receptor subfamily 4 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg19_v2_chr12_+_52445191_52445243 | 0.41 | 2.3e-02 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63638339 | 7.18 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr18_-_24722995 | 2.10 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr1_+_104293028 | 2.04 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr20_+_31755934 | 1.59 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr1_+_212738676 | 1.34 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr9_-_75567962 | 1.22 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr1_+_61869748 | 0.99 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chrX_-_108868390 | 0.92 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr16_+_53241854 | 0.83 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_1051481 | 0.82 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr11_+_7618413 | 0.80 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_+_73109339 | 0.79 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr3_+_184055240 | 0.78 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr1_+_16062820 | 0.77 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr22_+_24891210 | 0.75 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr2_-_25194963 | 0.75 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_-_105516923 | 0.72 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr12_+_109577202 | 0.72 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_+_13134772 | 0.70 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_-_152489484 | 0.66 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_111743285 | 0.65 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr4_+_71108300 | 0.64 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr8_+_99956759 | 0.64 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr19_+_14138960 | 0.62 |
ENST00000431365.2
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr16_+_691792 | 0.55 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr2_-_207024233 | 0.54 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_174769006 | 0.52 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_105517021 | 0.52 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr7_+_99933730 | 0.51 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr19_+_17416457 | 0.50 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr19_+_17416609 | 0.50 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr6_-_116381918 | 0.49 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr19_+_49713991 | 0.48 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr6_+_31895254 | 0.47 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr20_+_12989596 | 0.46 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr5_-_169626104 | 0.45 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr14_-_35591433 | 0.44 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr18_-_52969844 | 0.42 |
ENST00000561831.3
|
TCF4
|
transcription factor 4 |
| chr17_+_73629500 | 0.40 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr14_-_101295407 | 0.40 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr2_+_234216454 | 0.40 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr10_+_104005272 | 0.40 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr7_-_37488834 | 0.39 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_+_3876859 | 0.38 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr15_-_42783303 | 0.38 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr3_+_14989186 | 0.37 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr3_+_29322437 | 0.36 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr1_+_14075903 | 0.36 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr3_+_101443476 | 0.34 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr20_-_44485835 | 0.34 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr12_+_32655110 | 0.33 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_149068464 | 0.32 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr8_+_123793633 | 0.31 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr6_+_31895467 | 0.30 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr7_-_72936531 | 0.30 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr10_-_17171817 | 0.30 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr1_-_33815486 | 0.29 |
ENST00000373418.3
|
PHC2
|
polyhomeotic homolog 2 (Drosophila) |
| chr22_-_51016846 | 0.28 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr6_+_69942298 | 0.26 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr17_-_66287257 | 0.26 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr22_-_51017084 | 0.26 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr17_-_7123021 | 0.25 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr3_+_138340049 | 0.25 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_-_178257401 | 0.25 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr10_-_61666267 | 0.25 |
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6 |
| chr20_+_17680587 | 0.24 |
ENST00000427254.1
ENST00000377805.3 |
BANF2
|
barrier to autointegration factor 2 |
| chr14_-_77495007 | 0.24 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr2_-_25194476 | 0.24 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr8_+_99956662 | 0.23 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr4_-_74088800 | 0.22 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr12_+_49687425 | 0.22 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr20_+_11898507 | 0.22 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr5_-_74162605 | 0.22 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr3_-_44519131 | 0.21 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr1_+_40713573 | 0.21 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr3_+_14989076 | 0.21 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_-_16759711 | 0.21 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_+_64346386 | 0.20 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr12_+_54378923 | 0.20 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr6_-_33771757 | 0.20 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr6_-_152958521 | 0.19 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr20_+_17680599 | 0.19 |
ENST00000246090.5
|
BANF2
|
barrier to autointegration factor 2 |
| chr10_+_101542462 | 0.19 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr2_-_27603582 | 0.18 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr22_-_31364187 | 0.18 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr20_+_37590942 | 0.17 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr14_-_21567009 | 0.17 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr14_-_21566731 | 0.17 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr11_+_64073699 | 0.17 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr7_-_6048702 | 0.15 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr4_-_57253587 | 0.15 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr7_-_6048650 | 0.14 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr3_+_113465866 | 0.14 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr14_-_95623607 | 0.13 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr19_-_11639931 | 0.12 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr3_+_51741072 | 0.12 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr19_-_11639910 | 0.12 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr2_-_44223138 | 0.11 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr15_-_41120896 | 0.10 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr16_+_21312170 | 0.09 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr7_-_72936608 | 0.09 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr8_+_133787586 | 0.09 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr12_-_95467267 | 0.09 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr7_+_119913688 | 0.08 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr17_-_66287310 | 0.08 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr17_+_41150479 | 0.08 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr4_-_40632605 | 0.08 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr11_+_278365 | 0.08 |
ENST00000534750.1
|
NLRP6
|
NLR family, pyrin domain containing 6 |
| chr3_+_184038073 | 0.08 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr7_-_81399411 | 0.08 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_31895480 | 0.07 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr13_-_33112823 | 0.07 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr11_-_111649015 | 0.07 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr4_-_89744314 | 0.07 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr7_-_81399329 | 0.07 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_+_22386325 | 0.07 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr9_-_139137648 | 0.07 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr18_+_43753974 | 0.07 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr17_-_29624343 | 0.07 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr1_+_173793777 | 0.06 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr10_+_35484793 | 0.06 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr1_+_165600436 | 0.06 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr7_+_6048856 | 0.06 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr4_-_89744457 | 0.06 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_+_75335604 | 0.05 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr15_+_36887069 | 0.05 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr3_+_184037466 | 0.05 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr10_-_74714533 | 0.05 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chr17_+_41150793 | 0.04 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr19_+_6372444 | 0.04 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr17_+_41150290 | 0.04 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr17_-_2169425 | 0.02 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr4_-_89744365 | 0.02 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr9_+_12693336 | 0.02 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr1_-_197036364 | 0.00 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr4_+_100495864 | 0.00 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr18_-_60986962 | 0.00 |
ENST00000333681.4
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr17_+_7123125 | 0.00 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr19_+_4153598 | 0.00 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 1.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 1.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.9 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.2 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 1.6 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 1.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 1.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004530 | ribonuclease III activity(GO:0004525) deoxyribonuclease I activity(GO:0004530) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |