Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
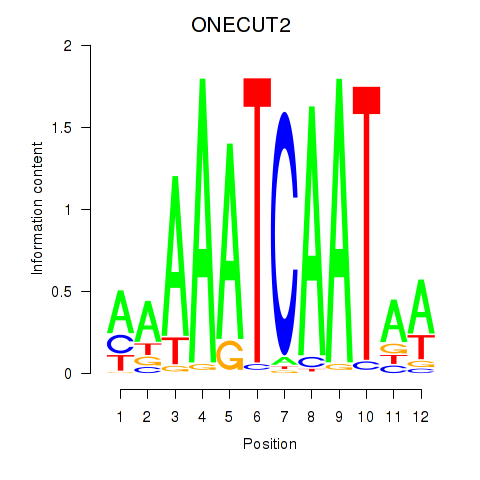
Results for ONECUT2_ONECUT3
Z-value: 0.78
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | 0.61 | 3.3e-04 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | 0.31 | 9.9e-02 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4385230 | 4.90 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr2_-_192016316 | 3.97 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chrX_+_100805496 | 2.19 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chr2_+_102413726 | 2.18 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_23510086 | 2.15 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr3_+_159557637 | 1.67 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr11_-_107729887 | 1.60 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_-_208030647 | 1.53 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_162887556 | 1.48 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr3_+_63428752 | 1.39 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr3_+_121774202 | 1.31 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr8_-_117043 | 1.31 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr14_+_37126765 | 1.25 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr5_+_140800638 | 1.17 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_+_367640 | 1.13 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr5_+_180794269 | 1.13 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor, family 4, subfamily F, member 3 |
| chr7_-_143059780 | 1.09 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr7_-_143059845 | 1.08 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr4_-_70826725 | 1.08 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr7_+_80231466 | 1.07 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr20_-_1309809 | 1.05 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr14_-_24047965 | 1.03 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr3_+_130650738 | 1.02 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_23012122 | 1.00 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr7_-_28220354 | 0.99 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr8_+_31497271 | 0.94 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr20_-_22566089 | 0.93 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr5_-_146833222 | 0.93 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr17_-_41623691 | 0.92 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr14_+_55034599 | 0.91 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr5_-_146833485 | 0.89 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr21_+_39628852 | 0.86 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_55646412 | 0.86 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr18_+_42260861 | 0.85 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr15_-_83837983 | 0.81 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr7_-_121944491 | 0.81 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr10_-_121296045 | 0.80 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr22_-_36220420 | 0.78 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_4535233 | 0.78 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr9_-_27529726 | 0.76 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr17_-_2304365 | 0.76 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr19_+_7598890 | 0.74 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr6_+_130339710 | 0.74 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_-_26233423 | 0.73 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr9_+_77230499 | 0.70 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr2_+_105050794 | 0.69 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr1_+_99127225 | 0.67 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr2_+_58655461 | 0.66 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr1_+_207943667 | 0.66 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr5_+_126984710 | 0.65 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr10_+_70980051 | 0.64 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr3_-_24207039 | 0.64 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr7_+_129906660 | 0.63 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr17_-_41623716 | 0.63 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr12_+_9144626 | 0.63 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr10_+_71561704 | 0.62 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr11_+_8040739 | 0.60 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr12_-_54653313 | 0.59 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr19_+_7599128 | 0.59 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr3_-_141747439 | 0.57 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_64225508 | 0.55 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr11_-_102651343 | 0.53 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr9_-_95244781 | 0.52 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr20_+_36405665 | 0.52 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr15_+_93443419 | 0.50 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr11_+_18154059 | 0.50 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chrX_-_64196376 | 0.49 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr3_+_63428982 | 0.48 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chrX_-_64196307 | 0.47 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_36338242 | 0.46 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr6_-_151773232 | 0.46 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr10_+_71561649 | 0.46 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chrX_-_64196351 | 0.45 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr4_-_20985632 | 0.45 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr15_-_99789736 | 0.44 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr5_+_138940742 | 0.41 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr1_-_622053 | 0.41 |
ENST00000332831.2
|
OR4F16
|
olfactory receptor, family 4, subfamily F, member 16 |
| chr19_-_52674896 | 0.41 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr17_-_202579 | 0.41 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr12_+_59989918 | 0.40 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_-_14026063 | 0.40 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr5_-_143550241 | 0.40 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr10_+_71562180 | 0.39 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_+_114178512 | 0.39 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr10_+_133753533 | 0.38 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr21_+_39628655 | 0.38 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_71587669 | 0.38 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_+_4699244 | 0.38 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_+_190448095 | 0.38 |
ENST00000424735.1
|
RP11-547I7.2
|
RP11-547I7.2 |
| chr1_-_243326612 | 0.37 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr2_+_241625749 | 0.36 |
ENST00000407635.2
|
AC011298.2
|
AC011298.2 |
| chr3_-_71632894 | 0.33 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr2_-_163695128 | 0.33 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr10_-_73975657 | 0.33 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr4_+_169418255 | 0.33 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_+_45596398 | 0.32 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr17_-_46691990 | 0.31 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr16_+_72459838 | 0.30 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr20_-_62587735 | 0.30 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr1_+_159557607 | 0.30 |
ENST00000255040.2
|
APCS
|
amyloid P component, serum |
| chr5_-_9630463 | 0.29 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr12_-_62586543 | 0.28 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr14_+_93357749 | 0.27 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr1_-_894620 | 0.27 |
ENST00000327044.6
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae) |
| chr14_+_37131058 | 0.27 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr12_+_100594557 | 0.26 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr4_+_174818390 | 0.26 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr3_+_52811596 | 0.26 |
ENST00000542827.1
ENST00000273283.2 |
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr10_-_98031265 | 0.26 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr4_+_71248795 | 0.26 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr19_-_44952635 | 0.25 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr3_+_188889737 | 0.25 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr4_+_187187098 | 0.25 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr5_-_143550159 | 0.24 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chrY_+_20137667 | 0.24 |
ENST00000250838.4
ENST00000426790.1 |
CDY2A
|
chromodomain protein, Y-linked, 2A |
| chrY_-_19992098 | 0.24 |
ENST00000544303.1
ENST00000382867.3 |
CDY2B
|
chromodomain protein, Y-linked, 2B |
| chrX_+_90689810 | 0.24 |
ENST00000312600.3
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5 |
| chr20_-_1974692 | 0.24 |
ENST00000217305.2
ENST00000539905.1 |
PDYN
|
prodynorphin |
| chr12_+_48866448 | 0.24 |
ENST00000266594.1
|
ANP32D
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member D |
| chr8_+_70850403 | 0.23 |
ENST00000602248.1
|
AC090574.1
|
Uncharacterized protein |
| chr2_-_50201327 | 0.23 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chrY_+_27768264 | 0.23 |
ENST00000361963.2
ENST00000306609.4 |
CDY1
|
chromodomain protein, Y-linked, 1 |
| chr11_-_116658758 | 0.23 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr6_+_29068386 | 0.22 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr16_-_20702578 | 0.22 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr17_-_46692287 | 0.21 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr2_+_67624430 | 0.21 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr5_-_135701164 | 0.21 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr3_-_187388173 | 0.21 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr16_-_69788816 | 0.20 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr1_-_100598444 | 0.20 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr4_+_71588372 | 0.20 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_+_56954808 | 0.20 |
ENST00000510483.1
ENST00000370706.4 ENST00000357489.3 |
ZNF451
|
zinc finger protein 451 |
| chr7_-_14026123 | 0.20 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr11_-_116658695 | 0.19 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr11_-_60623437 | 0.19 |
ENST00000332539.4
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr2_-_163695238 | 0.19 |
ENST00000328032.4
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr4_-_69536346 | 0.19 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_+_248201474 | 0.19 |
ENST00000366479.2
|
OR2L2
|
olfactory receptor, family 2, subfamily L, member 2 |
| chrX_+_123480194 | 0.18 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr4_+_156824840 | 0.18 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr6_+_27925019 | 0.18 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr4_-_103747011 | 0.17 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103746683 | 0.17 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_77593474 | 0.17 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr10_-_98031310 | 0.17 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr17_-_66951474 | 0.17 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr8_+_77593448 | 0.17 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr15_+_54901540 | 0.17 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr1_+_117297007 | 0.16 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr5_+_65440032 | 0.16 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr2_-_220174166 | 0.16 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr17_-_4167142 | 0.16 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr5_+_147774275 | 0.16 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_-_39348137 | 0.16 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_-_69434245 | 0.15 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chrY_-_26194116 | 0.15 |
ENST00000306882.4
ENST00000382407.1 |
CDY1B
|
chromodomain protein, Y-linked, 1B |
| chr17_-_79876010 | 0.15 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr2_+_7017796 | 0.15 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr1_-_111150048 | 0.15 |
ENST00000485317.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr3_-_141747459 | 0.14 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr20_+_43849941 | 0.14 |
ENST00000372769.3
|
SEMG2
|
semenogelin II |
| chr4_-_103746924 | 0.14 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_169418195 | 0.14 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_175839551 | 0.14 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr17_-_37764128 | 0.14 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr14_-_23426322 | 0.14 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr2_+_166095898 | 0.13 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr14_+_88490894 | 0.13 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr9_-_28670283 | 0.12 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr14_+_22265444 | 0.12 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr14_-_23426270 | 0.12 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr15_+_38226827 | 0.11 |
ENST00000559502.1
ENST00000558148.1 ENST00000558158.1 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr1_+_207262627 | 0.11 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr5_-_156390230 | 0.11 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr14_-_23426337 | 0.11 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr1_-_159684371 | 0.11 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr11_+_6947647 | 0.11 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr1_+_207262578 | 0.11 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr4_+_175839506 | 0.10 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr19_+_45596218 | 0.10 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr3_+_46283916 | 0.10 |
ENST00000395940.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr1_-_146068184 | 0.10 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr12_+_20848282 | 0.10 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_20848377 | 0.10 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr3_-_186080012 | 0.09 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr1_+_207262540 | 0.09 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr4_-_102268484 | 0.09 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_115259337 | 0.09 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr7_+_120702819 | 0.09 |
ENST00000423795.1
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_+_71122974 | 0.08 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr4_-_111563076 | 0.08 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr9_+_33629119 | 0.08 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr2_+_171034646 | 0.08 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr16_-_2013101 | 0.08 |
ENST00000526586.2
|
RPS2
|
ribosomal protein S2 |
| chr1_-_46664074 | 0.08 |
ENST00000371986.3
ENST00000371984.3 |
POMGNT1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr4_+_187187337 | 0.07 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr7_+_129007964 | 0.07 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_+_144811744 | 0.07 |
ENST00000338347.4
ENST00000440491.2 ENST00000375552.4 |
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chrX_-_19988382 | 0.07 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.4 | 1.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.3 | 0.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 4.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 1.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.9 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 2.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.7 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.5 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 3.7 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 1.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.6 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 2.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.9 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.0 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 1.5 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.7 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 2.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.2 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |