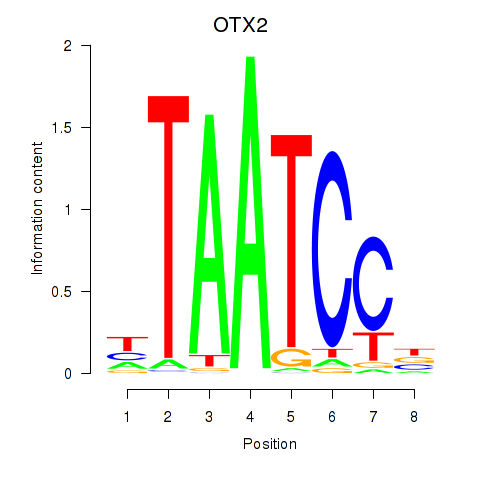
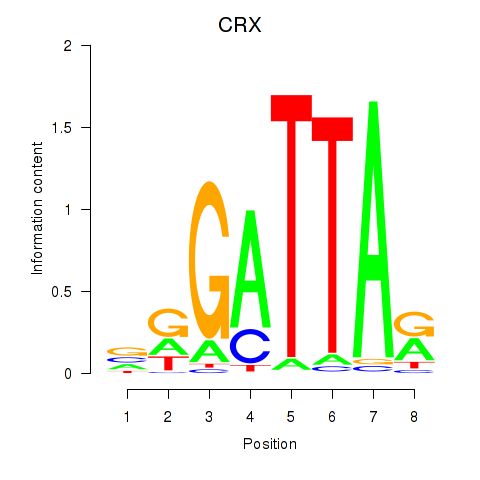
Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for OTX2_CRX
Z-value: 0.67
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX2 | hg19_v2_chr14_-_57277178_57277203 | 0.66 | 7.5e-05 | Click! |
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.41 | 2.3e-02 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_38859996 | 9.62 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr8_+_54764346 | 3.83 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr15_-_74495188 | 2.63 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr19_-_51538118 | 2.49 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_-_51538148 | 2.47 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr6_+_130339710 | 2.14 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_-_149095652 | 1.80 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr9_+_75229616 | 1.76 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chrX_+_69488174 | 1.72 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr5_-_149669192 | 1.66 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_+_116298778 | 1.53 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr8_-_91095099 | 1.44 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr16_+_58010339 | 1.40 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr2_+_219472488 | 1.39 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chr18_-_28681950 | 1.34 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr1_+_180897269 | 1.32 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chrX_+_69488155 | 1.28 |
ENST00000374495.3
|
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr19_+_46001697 | 1.18 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chrX_+_69509927 | 1.18 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr15_+_33603147 | 1.09 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr18_-_64271316 | 1.08 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr12_-_2027639 | 1.04 |
ENST00000586184.1
ENST00000587995.1 ENST00000585732.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr11_-_62752162 | 1.01 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr6_-_35480640 | 1.00 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr18_-_28682374 | 0.96 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr9_-_39288092 | 0.94 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr12_+_53693466 | 0.91 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr1_-_150208320 | 0.85 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_+_53693812 | 0.82 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr1_+_55505184 | 0.81 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr10_-_105845674 | 0.79 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr1_+_11751748 | 0.78 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr3_+_111393501 | 0.78 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr8_+_70378852 | 0.75 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr15_-_34610962 | 0.74 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr12_-_53189892 | 0.73 |
ENST00000309505.3
ENST00000417996.2 |
KRT3
|
keratin 3 |
| chr16_-_58004992 | 0.72 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr12_-_53171128 | 0.72 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chrX_+_135279179 | 0.72 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr19_-_51537982 | 0.70 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr11_+_20044096 | 0.67 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr11_+_124735282 | 0.65 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr6_+_34433844 | 0.64 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr6_+_35310312 | 0.62 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr3_-_54962100 | 0.62 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr17_-_43209862 | 0.62 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr12_+_7941989 | 0.61 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr19_+_48325097 | 0.60 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr17_-_60885659 | 0.60 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chrX_+_41306575 | 0.60 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr17_-_60885700 | 0.59 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chrX_+_135278908 | 0.58 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr6_-_66417107 | 0.57 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr6_+_35310391 | 0.57 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr7_-_83824169 | 0.56 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_+_18474204 | 0.55 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr7_+_93535817 | 0.54 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr13_-_36050819 | 0.54 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr3_+_130650738 | 0.54 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_-_86001210 | 0.54 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr20_+_42984330 | 0.54 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr12_-_54779511 | 0.51 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_-_47950745 | 0.51 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chrX_-_124097620 | 0.50 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr9_+_34179003 | 0.50 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr10_-_48390973 | 0.50 |
ENST00000224600.4
|
RBP3
|
retinol binding protein 3, interstitial |
| chr5_-_95768973 | 0.50 |
ENST00000311106.3
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1 |
| chr11_-_8680383 | 0.49 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr3_-_48229846 | 0.49 |
ENST00000302506.3
ENST00000351231.3 ENST00000437972.1 |
CDC25A
|
cell division cycle 25A |
| chr1_-_231560790 | 0.48 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr1_+_159409512 | 0.48 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr14_-_95236551 | 0.47 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr6_-_30658745 | 0.45 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr17_+_43318434 | 0.45 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr14_+_96722152 | 0.44 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr14_-_58894332 | 0.44 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr11_-_62752455 | 0.43 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr2_+_219472637 | 0.42 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr7_+_80231466 | 0.42 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr18_-_64271363 | 0.42 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr1_+_62439037 | 0.42 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr19_-_42931567 | 0.42 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr18_+_616672 | 0.42 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr9_-_95244781 | 0.40 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr5_-_41510656 | 0.40 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_-_150208363 | 0.40 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208291 | 0.40 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_-_41510725 | 0.39 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr7_+_28452130 | 0.39 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr7_+_93535866 | 0.39 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr18_+_32290218 | 0.39 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr3_+_50649302 | 0.38 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_+_85822839 | 0.38 |
ENST00000441634.1
|
RNF181
|
ring finger protein 181 |
| chr11_-_111783595 | 0.38 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_145718587 | 0.38 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr6_-_138893661 | 0.37 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr11_-_62752429 | 0.36 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_-_11589131 | 0.36 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr8_-_25281747 | 0.35 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr14_-_22005062 | 0.35 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_39395165 | 0.35 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr11_+_72975524 | 0.34 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr18_+_616711 | 0.34 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_+_72975578 | 0.34 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_-_71640227 | 0.33 |
ENST00000388726.3
ENST00000392650.3 |
SDK2
|
sidekick cell adhesion molecule 2 |
| chr9_+_103204553 | 0.33 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr11_+_72975559 | 0.33 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr2_+_192141611 | 0.33 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr11_+_128563652 | 0.33 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_-_153601136 | 0.32 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr15_+_93443419 | 0.32 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr6_-_88411911 | 0.31 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr2_-_119605253 | 0.31 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr5_-_11588907 | 0.31 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr16_+_8715536 | 0.31 |
ENST00000563958.1
ENST00000381920.3 ENST00000564554.1 |
METTL22
|
methyltransferase like 22 |
| chr18_-_35145728 | 0.31 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr9_-_131486367 | 0.30 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr2_+_120770581 | 0.29 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr9_+_18474098 | 0.29 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr1_+_153651078 | 0.29 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr3_-_50649192 | 0.29 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr8_-_80993010 | 0.28 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr17_+_7905912 | 0.28 |
ENST00000254854.4
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific) |
| chr12_+_81101277 | 0.28 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr16_+_81272287 | 0.28 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_-_70780572 | 0.28 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr2_+_201170703 | 0.28 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_+_79675965 | 0.28 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr10_-_76868931 | 0.27 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr15_+_58430567 | 0.26 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr20_+_59654146 | 0.26 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr19_-_48547294 | 0.26 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr3_-_137851220 | 0.25 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr19_+_56166360 | 0.24 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr1_-_150208412 | 0.24 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_+_28605516 | 0.24 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr11_-_36619771 | 0.24 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr15_+_58430368 | 0.24 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr6_+_31555045 | 0.23 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr10_+_52751010 | 0.23 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr13_-_33780133 | 0.22 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr19_+_39989580 | 0.22 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr11_-_75017734 | 0.22 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chrX_-_153363125 | 0.22 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chrY_+_16634483 | 0.22 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr1_-_53608289 | 0.22 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr3_-_98241358 | 0.21 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_-_52604607 | 0.21 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr19_+_39989535 | 0.21 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr20_-_31124186 | 0.21 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr10_+_24528108 | 0.21 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr2_-_89459813 | 0.21 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr10_+_102505468 | 0.21 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr8_-_133123406 | 0.21 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr2_+_201170596 | 0.20 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr10_+_103986085 | 0.20 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr1_-_11751665 | 0.20 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr18_-_70532906 | 0.20 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_-_11751529 | 0.20 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr5_-_36301984 | 0.19 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr10_+_95372289 | 0.19 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chrX_-_21676442 | 0.19 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr17_-_39661849 | 0.19 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr1_+_81771806 | 0.18 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_+_144989309 | 0.18 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr15_-_65407524 | 0.18 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr2_+_33661382 | 0.18 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_173174681 | 0.18 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr3_+_130279178 | 0.18 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr11_-_111794446 | 0.17 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr19_-_3479086 | 0.17 |
ENST00000587847.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chrX_-_54384425 | 0.17 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr12_-_14849470 | 0.17 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr15_-_31453157 | 0.16 |
ENST00000559177.1
ENST00000558445.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr21_-_35016231 | 0.16 |
ENST00000438788.1
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr18_-_21891460 | 0.16 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr1_-_150208498 | 0.16 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr8_+_42873548 | 0.16 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr9_+_18474163 | 0.16 |
ENST00000380566.4
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr2_+_85822857 | 0.16 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr12_-_110937351 | 0.16 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr2_-_14541060 | 0.15 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr7_-_129592700 | 0.15 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr2_-_55237484 | 0.15 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr1_-_53608249 | 0.15 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr8_+_65492756 | 0.15 |
ENST00000321870.1
|
BHLHE22
|
basic helix-loop-helix family, member e22 |
| chr17_+_48911942 | 0.14 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chrX_-_119077695 | 0.14 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr5_-_524445 | 0.14 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr2_+_120770645 | 0.14 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_115219171 | 0.14 |
ENST00000409163.1
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr6_+_155537771 | 0.14 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr12_-_67197760 | 0.14 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr2_-_30144432 | 0.13 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr5_-_77072085 | 0.13 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr1_+_244816237 | 0.13 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr6_-_30899924 | 0.13 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr11_-_67290867 | 0.13 |
ENST00000353903.5
ENST00000294288.4 |
CABP2
|
calcium binding protein 2 |
| chr1_-_102312517 | 0.13 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr18_+_29769978 | 0.13 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.5 | 2.6 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 0.8 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 2.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 0.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 1.6 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.2 | 2.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.7 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 16.9 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 2.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 1.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 1.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.5 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 1.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.6 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 1.7 | GO:0038166 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.6 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.4 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.2 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 2.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 1.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 1.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 1.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0060745 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) mammary gland branching involved in pregnancy(GO:0060745) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 1.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 2.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 4.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 11.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 1.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 0.8 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 2.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 0.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.0 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.5 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 2.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.2 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 2.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 6.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 4.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |