Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PAX1_PAX9
Z-value: 0.59
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
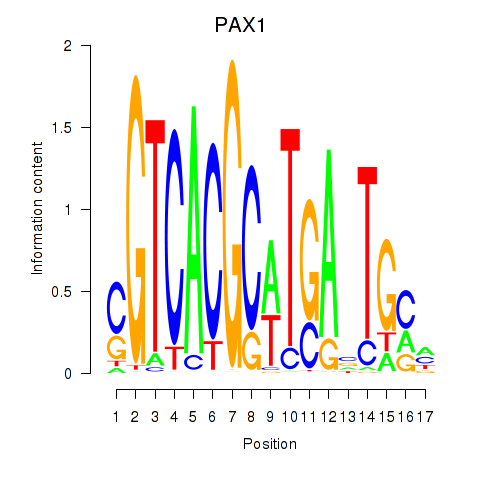
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
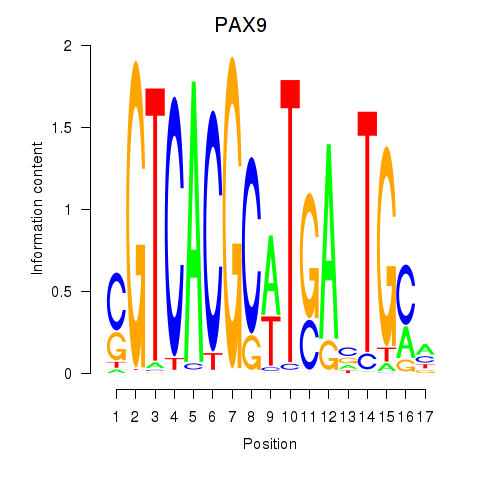
PAX9
|
ENSG00000198807.8 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37126765_37126799 | 0.39 | 3.3e-02 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | 0.21 | 2.6e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23261589 | 4.64 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr4_+_75230853 | 4.43 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr7_+_36429424 | 2.40 |
ENST00000396068.2
|
ANLN
|
anillin, actin binding protein |
| chr7_+_36429409 | 2.35 |
ENST00000265748.2
|
ANLN
|
anillin, actin binding protein |
| chr9_+_33750667 | 2.31 |
ENST00000457896.1
ENST00000342836.4 ENST00000429677.3 |
PRSS3
|
protease, serine, 3 |
| chr9_+_33750515 | 2.31 |
ENST00000361005.5
|
PRSS3
|
protease, serine, 3 |
| chr9_+_33795533 | 1.72 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr11_+_12308447 | 1.38 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr19_-_40791211 | 1.23 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr18_-_33077556 | 1.20 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr13_+_98086445 | 0.82 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr7_+_142457315 | 0.80 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr17_-_33814851 | 0.80 |
ENST00000449046.1
ENST00000260908.7 |
SLFN12L
|
schlafen family member 12-like |
| chr17_+_39969183 | 0.73 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr17_+_15848231 | 0.58 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr9_-_124991124 | 0.57 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr8_-_125384927 | 0.57 |
ENST00000297632.6
|
TMEM65
|
transmembrane protein 65 |
| chr6_-_137494775 | 0.47 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chr17_-_7137582 | 0.44 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr5_+_72251857 | 0.43 |
ENST00000507345.2
ENST00000512348.1 ENST00000287761.6 |
FCHO2
|
FCH domain only 2 |
| chr8_-_101965559 | 0.42 |
ENST00000353245.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_137366163 | 0.38 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr5_-_133747589 | 0.34 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr19_-_40791302 | 0.33 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr14_-_69261310 | 0.31 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr2_+_234160217 | 0.30 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr1_-_204165610 | 0.29 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr2_+_234160340 | 0.26 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr11_+_6947720 | 0.26 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr7_-_144533074 | 0.26 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr7_-_112758665 | 0.25 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_-_112758589 | 0.25 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr17_-_39968406 | 0.24 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr19_+_14184370 | 0.23 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr3_-_57113314 | 0.23 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr2_+_119699864 | 0.23 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr4_+_123747834 | 0.23 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr10_-_99393242 | 0.21 |
ENST00000370635.3
ENST00000478953.1 ENST00000335628.3 |
MORN4
|
MORN repeat containing 4 |
| chr11_+_6947647 | 0.20 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr14_+_64854958 | 0.20 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr9_-_104249400 | 0.19 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr4_-_171011084 | 0.17 |
ENST00000337664.4
|
AADAT
|
aminoadipate aminotransferase |
| chr17_-_39968855 | 0.16 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr19_-_409134 | 0.16 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr4_-_171011323 | 0.15 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr15_+_63569785 | 0.14 |
ENST00000380343.4
ENST00000560353.1 |
APH1B
|
APH1B gamma secretase subunit |
| chr8_-_101965146 | 0.14 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_-_77997126 | 0.13 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr10_-_99393208 | 0.13 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr10_-_43904608 | 0.13 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_+_57828521 | 0.12 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr17_+_37356555 | 0.12 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr17_+_37356586 | 0.11 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr1_+_26758790 | 0.11 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr7_-_19157248 | 0.11 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr17_+_37356528 | 0.10 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr11_+_63870660 | 0.05 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr6_+_159291090 | 0.05 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr9_-_21142144 | 0.05 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr1_+_26759295 | 0.05 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr6_+_34857019 | 0.03 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr4_-_163085141 | 0.03 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr4_+_123747979 | 0.03 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr18_+_39766626 | 0.02 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr14_+_39703112 | 0.02 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr6_+_159290917 | 0.02 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr8_+_39792474 | 0.02 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr1_+_45140360 | 0.01 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 1.5 | 4.4 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.4 | 1.6 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.2 | 7.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.7 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.6 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 4.6 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.3 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.6 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.8 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.3 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 6.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 4.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 4.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 6.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 11.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |