Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
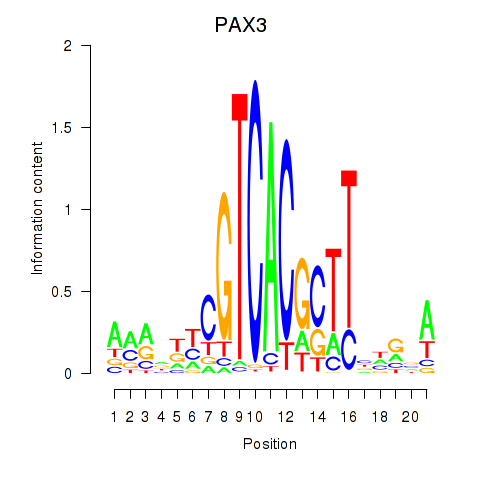
Results for PAX3
Z-value: 0.72
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.14 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg19_v2_chr2_-_223163465_223163730 | 0.15 | 4.4e-01 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_38420783 | 2.39 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chrX_+_38420623 | 2.27 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr6_-_4135693 | 1.75 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr15_-_80263506 | 1.51 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr15_+_45722727 | 1.31 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr13_+_96085847 | 1.29 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr9_+_33795533 | 1.06 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr10_-_106098162 | 0.99 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr4_-_104119528 | 0.90 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr5_+_7654057 | 0.88 |
ENST00000537121.1
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr4_+_156680143 | 0.76 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr7_-_148580563 | 0.75 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr8_-_10512569 | 0.75 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr3_+_160117087 | 0.73 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr7_+_142031986 | 0.71 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr11_+_313503 | 0.63 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr9_-_132515302 | 0.59 |
ENST00000340607.4
|
PTGES
|
prostaglandin E synthase |
| chr19_-_2783363 | 0.59 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr19_-_48048518 | 0.59 |
ENST00000595558.1
ENST00000263351.5 |
ZNF541
|
zinc finger protein 541 |
| chr6_-_9939552 | 0.58 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr17_-_38574169 | 0.57 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr7_-_30066233 | 0.56 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr4_+_156680153 | 0.55 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chrX_+_103217207 | 0.54 |
ENST00000563257.1
ENST00000540220.1 ENST00000436583.1 ENST00000567181.1 ENST00000569577.1 |
TMSB15B
|
thymosin beta 15B |
| chr7_-_19813192 | 0.54 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr19_+_4343584 | 0.50 |
ENST00000596722.1
|
MPND
|
MPN domain containing |
| chr9_+_131549483 | 0.50 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr5_+_70220768 | 0.49 |
ENST00000351205.4
ENST00000503079.2 ENST00000380707.4 ENST00000514951.1 ENST00000506163.1 |
SMN1
|
survival of motor neuron 1, telomeric |
| chr4_-_168155730 | 0.48 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr22_+_20105259 | 0.44 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr3_+_160117418 | 0.43 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr4_-_168155700 | 0.43 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_+_69345350 | 0.40 |
ENST00000380741.4
ENST00000380743.4 ENST00000511812.1 ENST00000380742.4 |
SMN2
|
survival of motor neuron 2, centromeric |
| chr14_+_22964877 | 0.40 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr11_+_66624527 | 0.39 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr15_+_99791716 | 0.39 |
ENST00000558172.1
ENST00000561276.1 ENST00000331450.5 |
LRRC28
|
leucine rich repeat containing 28 |
| chr19_+_7049332 | 0.39 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
| chrX_-_53461288 | 0.38 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr20_-_44539538 | 0.38 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr9_+_131549610 | 0.37 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr10_-_126432821 | 0.37 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr2_+_240323439 | 0.36 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr2_+_44589036 | 0.36 |
ENST00000402247.1
ENST00000407131.1 ENST00000403853.3 ENST00000378494.3 |
CAMKMT
|
calmodulin-lysine N-methyltransferase |
| chr14_+_20937538 | 0.36 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chrX_+_43515467 | 0.36 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chrX_-_15872914 | 0.35 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr18_-_59274139 | 0.34 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr19_+_55851221 | 0.34 |
ENST00000255613.3
ENST00000539076.1 |
SUV420H2
AC020922.1
|
suppressor of variegation 4-20 homolog 2 (Drosophila) Uncharacterized protein |
| chr22_-_21581926 | 0.33 |
ENST00000401924.1
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr8_+_92082424 | 0.33 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chrX_-_53461305 | 0.33 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_+_12524965 | 0.32 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chrX_+_135230712 | 0.32 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_84630053 | 0.32 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_168155577 | 0.32 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_112709804 | 0.32 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr21_+_34775181 | 0.32 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr10_-_75226166 | 0.32 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_-_119379719 | 0.32 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr9_-_13175823 | 0.31 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chrX_+_102883620 | 0.31 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr8_-_145159083 | 0.31 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr5_-_157079428 | 0.30 |
ENST00000265007.6
|
SOX30
|
SRY (sex determining region Y)-box 30 |
| chr6_+_31939608 | 0.30 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chrX_+_153770421 | 0.29 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr10_+_134150835 | 0.29 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr4_-_168155417 | 0.29 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr8_-_81083731 | 0.28 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr17_+_35732955 | 0.28 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr17_+_35732916 | 0.28 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr3_+_112709755 | 0.28 |
ENST00000383678.2
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr22_+_42372764 | 0.27 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr19_+_4343691 | 0.26 |
ENST00000597036.1
|
MPND
|
MPN domain containing |
| chr12_+_10460549 | 0.26 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chrX_-_48931648 | 0.25 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr12_+_112563303 | 0.25 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr2_-_68479614 | 0.25 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr12_-_69080590 | 0.24 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr1_-_153123345 | 0.24 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr12_+_69080734 | 0.23 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr12_-_58131931 | 0.23 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_+_112563335 | 0.22 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr1_+_101185290 | 0.22 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr2_-_44065889 | 0.21 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr2_-_217724767 | 0.21 |
ENST00000236979.2
|
TNP1
|
transition protein 1 (during histone to protamine replacement) |
| chr3_+_49726932 | 0.21 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr2_-_128051670 | 0.21 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr19_-_42463418 | 0.21 |
ENST00000600292.1
ENST00000601078.1 ENST00000601891.1 ENST00000222008.6 |
RABAC1
|
Rab acceptor 1 (prenylated) |
| chr15_+_81591757 | 0.20 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr14_-_64804814 | 0.20 |
ENST00000554572.1
ENST00000358599.5 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr4_+_128982490 | 0.19 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr7_+_128399002 | 0.19 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr11_+_112038088 | 0.19 |
ENST00000530752.1
ENST00000280358.4 |
TEX12
|
testis expressed 12 |
| chr1_-_7913089 | 0.18 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr2_-_128051708 | 0.18 |
ENST00000285398.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr6_+_127587755 | 0.18 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr13_+_76445187 | 0.17 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr11_+_65122216 | 0.16 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr12_-_10562356 | 0.16 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr17_+_3118915 | 0.16 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr1_-_153931052 | 0.15 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr13_+_98605902 | 0.15 |
ENST00000460070.1
ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5
|
importin 5 |
| chr19_+_38085768 | 0.14 |
ENST00000316433.4
ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540
|
zinc finger protein 540 |
| chr12_+_10460417 | 0.14 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr10_+_96162242 | 0.14 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr7_+_142457315 | 0.14 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr16_+_23847339 | 0.14 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr17_+_32597232 | 0.14 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr9_-_114521783 | 0.14 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr11_-_19082216 | 0.13 |
ENST00000329773.2
|
MRGPRX2
|
MAS-related GPR, member X2 |
| chr15_+_99791567 | 0.13 |
ENST00000558879.1
ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28
|
leucine rich repeat containing 28 |
| chr2_+_234627424 | 0.13 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr15_+_38226827 | 0.12 |
ENST00000559502.1
ENST00000558148.1 ENST00000558158.1 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr2_-_114514181 | 0.12 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr2_-_218706877 | 0.12 |
ENST00000446688.1
|
TNS1
|
tensin 1 |
| chr3_-_119379427 | 0.12 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr9_-_73029540 | 0.11 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr17_+_37356555 | 0.11 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr3_-_51813009 | 0.11 |
ENST00000398780.3
|
IQCF6
|
IQ motif containing F6 |
| chr11_+_33563821 | 0.11 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr1_+_152758690 | 0.11 |
ENST00000368771.1
ENST00000368770.3 |
LCE1E
|
late cornified envelope 1E |
| chr2_+_183982238 | 0.11 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr17_+_37356586 | 0.11 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr17_+_37356528 | 0.11 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr14_+_22217447 | 0.11 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr17_-_3461092 | 0.10 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr19_-_46296011 | 0.10 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr14_+_22675388 | 0.10 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr1_+_171283331 | 0.10 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr5_-_87516448 | 0.10 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr12_-_57352103 | 0.09 |
ENST00000398138.3
|
RDH16
|
retinol dehydrogenase 16 (all-trans) |
| chr2_+_242811874 | 0.09 |
ENST00000343216.3
|
CXXC11
|
CXXC finger protein 11 |
| chr12_-_127544894 | 0.09 |
ENST00000546062.1
ENST00000512624.2 ENST00000540244.1 |
RP11-575F12.1
|
RP11-575F12.1 |
| chr7_+_99816859 | 0.09 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr17_-_59668550 | 0.09 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr21_-_37451680 | 0.09 |
ENST00000399201.1
|
SETD4
|
SET domain containing 4 |
| chr6_+_1080164 | 0.08 |
ENST00000314040.1
|
AL033381.1
|
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr13_+_53191605 | 0.08 |
ENST00000342657.3
ENST00000398039.1 |
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr2_+_152214098 | 0.08 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr1_-_24151903 | 0.08 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr22_+_39101728 | 0.08 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr19_-_54784353 | 0.08 |
ENST00000391746.1
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr1_-_24151892 | 0.07 |
ENST00000235958.4
|
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr6_+_14117872 | 0.07 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr13_+_113623509 | 0.07 |
ENST00000535094.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr17_-_39280419 | 0.07 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr1_+_158323244 | 0.07 |
ENST00000434258.1
|
CD1E
|
CD1e molecule |
| chr17_-_7835228 | 0.07 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr1_+_40505891 | 0.07 |
ENST00000372797.3
ENST00000372802.1 ENST00000449311.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr7_-_50633078 | 0.07 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr19_+_38085731 | 0.06 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr2_-_183106641 | 0.06 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_+_48398053 | 0.06 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr19_-_41903161 | 0.06 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr3_+_184018352 | 0.06 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr5_+_49961727 | 0.06 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_25194476 | 0.05 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr8_+_107670064 | 0.05 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr17_+_4855053 | 0.05 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chrX_+_47077632 | 0.05 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr11_-_104972158 | 0.05 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr16_-_75018968 | 0.05 |
ENST00000262144.6
|
WDR59
|
WD repeat domain 59 |
| chr12_+_9142131 | 0.05 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr12_+_7060432 | 0.05 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_128982416 | 0.05 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr12_-_56123444 | 0.05 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr19_+_55105085 | 0.05 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr4_+_15376165 | 0.04 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr13_-_81801115 | 0.04 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr7_+_116502527 | 0.04 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chrX_+_70364667 | 0.04 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr3_+_14716606 | 0.04 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr6_-_137494775 | 0.04 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chr14_+_23938891 | 0.03 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr6_+_127588020 | 0.03 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr1_-_154127518 | 0.03 |
ENST00000368559.3
ENST00000271854.3 |
NUP210L
|
nucleoporin 210kDa-like |
| chr4_+_14113592 | 0.03 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr1_+_158323486 | 0.03 |
ENST00000444681.2
ENST00000368167.3 |
CD1E
|
CD1e molecule |
| chr11_+_9482512 | 0.02 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr6_-_24721054 | 0.02 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr1_+_26496362 | 0.02 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr1_+_247670415 | 0.02 |
ENST00000366491.2
ENST00000366489.1 ENST00000526896.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chrX_-_118699325 | 0.02 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr7_-_151574191 | 0.02 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr15_-_52861323 | 0.02 |
ENST00000569723.1
ENST00000249822.4 ENST00000567669.1 ENST00000569281.2 ENST00000563566.1 ENST00000567830.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr14_-_24711806 | 0.02 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_-_55001115 | 0.02 |
ENST00000493075.1
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr1_-_155214436 | 0.01 |
ENST00000327247.5
|
GBA
|
glucosidase, beta, acid |
| chrX_+_102883887 | 0.01 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr20_-_17511962 | 0.01 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chrY_-_20935572 | 0.01 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr15_+_51973550 | 0.01 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr11_+_9482551 | 0.01 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr15_+_51973680 | 0.01 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr3_+_183892635 | 0.01 |
ENST00000427072.1
ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr1_+_154244987 | 0.01 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr17_+_7835419 | 0.01 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr14_+_21510385 | 0.01 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr14_-_24584138 | 0.01 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr12_+_8309630 | 0.01 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 0.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.7 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.2 | 0.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 1.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.4 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.2 | GO:0060752 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.1 | 0.9 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |