Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
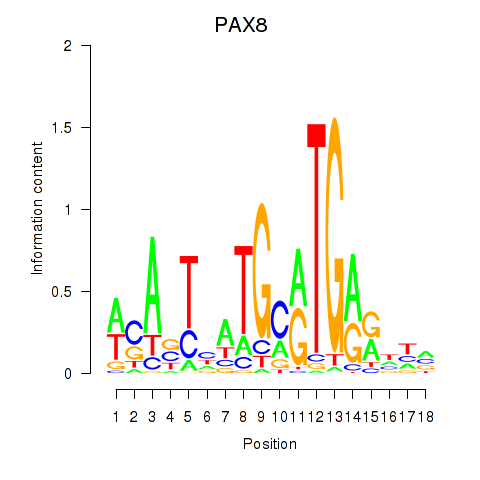
Results for PAX8
Z-value: 0.39
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_113999260_113999274 | 0.31 | 9.2e-02 | Click! |
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51487282 | 0.97 |
ENST00000595820.1
ENST00000597707.1 ENST00000336317.4 |
KLK7
|
kallikrein-related peptidase 7 |
| chr4_+_75230853 | 0.70 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr19_-_36019123 | 0.67 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_-_153013588 | 0.64 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr19_-_51537982 | 0.55 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_-_51538118 | 0.52 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_-_51538148 | 0.51 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr7_-_97501706 | 0.48 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr6_+_31126291 | 0.44 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr2_-_234763147 | 0.43 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr7_-_76255444 | 0.39 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr11_-_58343319 | 0.38 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr5_+_148521136 | 0.37 |
ENST00000506113.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr19_-_35992780 | 0.36 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr22_-_37403839 | 0.36 |
ENST00000402860.3
ENST00000381821.1 |
TEX33
|
testis expressed 33 |
| chr22_-_37403858 | 0.35 |
ENST00000405091.2
|
TEX33
|
testis expressed 33 |
| chr12_+_75874460 | 0.33 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_-_6961050 | 0.32 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr12_-_6960407 | 0.31 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr19_+_42301079 | 0.30 |
ENST00000596544.1
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr10_+_135340859 | 0.30 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr17_-_39216344 | 0.29 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr1_+_13359819 | 0.28 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr11_-_2170786 | 0.28 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr19_+_35630926 | 0.28 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_214638146 | 0.28 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr1_+_155006300 | 0.27 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr12_+_72058130 | 0.26 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr2_+_62900986 | 0.26 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr1_+_182419261 | 0.26 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr18_-_58040000 | 0.25 |
ENST00000299766.3
|
MC4R
|
melanocortin 4 receptor |
| chr1_-_119682812 | 0.24 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr3_+_121796697 | 0.23 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr8_+_27168988 | 0.23 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_-_186649543 | 0.23 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_54652060 | 0.23 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr16_-_28518153 | 0.23 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr1_+_87012753 | 0.22 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr2_-_37068530 | 0.22 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr6_+_43739697 | 0.21 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr9_-_116837249 | 0.21 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr1_+_16083154 | 0.20 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr8_+_31496809 | 0.19 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr4_+_156680153 | 0.19 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr11_-_105010320 | 0.19 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr11_+_117857063 | 0.18 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr11_+_5617858 | 0.18 |
ENST00000380097.3
|
TRIM6
|
tripartite motif containing 6 |
| chr12_-_53074182 | 0.18 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr15_+_67420441 | 0.18 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr10_+_24738355 | 0.18 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr6_-_32784687 | 0.18 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr17_+_80332153 | 0.18 |
ENST00000313135.2
|
UTS2R
|
urotensin 2 receptor |
| chr4_+_156680143 | 0.18 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr11_+_12766583 | 0.17 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr7_-_99277610 | 0.17 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr12_+_26348582 | 0.16 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr1_-_119683251 | 0.16 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr7_+_129984630 | 0.16 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr6_+_160542821 | 0.16 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chrX_-_71458802 | 0.16 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr17_-_74023291 | 0.16 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr2_+_17721230 | 0.16 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr8_+_59323823 | 0.15 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr9_-_21228221 | 0.15 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr8_+_39792474 | 0.14 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chrX_+_129535937 | 0.14 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr15_+_89631647 | 0.14 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr6_+_72926145 | 0.14 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr18_-_29264467 | 0.14 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr14_-_106610852 | 0.14 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr9_+_706842 | 0.14 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr12_+_57828521 | 0.14 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr4_+_118955500 | 0.14 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_-_21059029 | 0.13 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr1_-_182573514 | 0.13 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr19_+_50169081 | 0.13 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr6_+_160542870 | 0.13 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr3_-_137851220 | 0.13 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr17_+_67410832 | 0.13 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_+_18891045 | 0.13 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr2_-_136288113 | 0.13 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr7_-_144533074 | 0.13 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr12_-_56236734 | 0.12 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr19_+_50169216 | 0.12 |
ENST00000594157.1
ENST00000600947.1 ENST00000598306.1 |
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr6_+_14117872 | 0.12 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr7_+_37960163 | 0.12 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr1_-_222763240 | 0.12 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr4_+_80748619 | 0.12 |
ENST00000504263.1
|
PCAT4
|
prostate cancer associated transcript 4 (non-protein coding) |
| chr2_-_26864228 | 0.11 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr1_-_153514241 | 0.11 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr12_-_129308487 | 0.11 |
ENST00000266771.5
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr2_+_90248739 | 0.11 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr19_+_42300548 | 0.11 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr1_+_197382957 | 0.11 |
ENST00000367397.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr4_+_71600144 | 0.10 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_111852128 | 0.10 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr17_-_3499125 | 0.10 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr3_+_11178779 | 0.10 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr12_-_86650045 | 0.10 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_+_72922590 | 0.10 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr9_-_21239978 | 0.10 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr13_+_31309645 | 0.10 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr6_+_63921399 | 0.10 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chrX_+_151903207 | 0.10 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1 |
| chrX_+_99839799 | 0.10 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chrX_-_101410762 | 0.09 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr1_-_184006829 | 0.09 |
ENST00000361927.4
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr1_-_173176452 | 0.09 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr6_+_72922505 | 0.09 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr4_-_89951028 | 0.09 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr12_+_128399965 | 0.09 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr2_+_185463093 | 0.09 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr6_+_24126350 | 0.09 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr16_-_20367584 | 0.09 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr12_-_56236690 | 0.09 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr22_+_22730353 | 0.09 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr8_-_117043 | 0.09 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr2_+_171036635 | 0.09 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr8_-_66474884 | 0.09 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr8_+_49984894 | 0.09 |
ENST00000522267.1
ENST00000399653.4 ENST00000303202.8 |
C8orf22
|
chromosome 8 open reading frame 22 |
| chr9_-_128246769 | 0.08 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr9_+_27109133 | 0.08 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr18_-_29264669 | 0.08 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chrX_-_153141302 | 0.08 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr6_+_117198400 | 0.08 |
ENST00000332958.2
|
RFX6
|
regulatory factor X, 6 |
| chr9_+_118950325 | 0.08 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr15_-_43785274 | 0.08 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr17_+_1933404 | 0.08 |
ENST00000263083.6
ENST00000571418.1 |
DPH1
|
diphthamide biosynthesis 1 |
| chr19_+_12862604 | 0.08 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr4_-_80329356 | 0.08 |
ENST00000358842.3
|
GK2
|
glycerol kinase 2 |
| chr22_+_38082330 | 0.08 |
ENST00000359114.4
|
NOL12
|
nucleolar protein 12 |
| chr2_+_90198535 | 0.08 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr12_-_57410304 | 0.08 |
ENST00000441881.1
ENST00000458521.2 |
TAC3
|
tachykinin 3 |
| chr11_-_74022658 | 0.08 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr1_+_154300217 | 0.08 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr13_-_99174252 | 0.08 |
ENST00000376547.3
|
STK24
|
serine/threonine kinase 24 |
| chr7_+_7606497 | 0.08 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr21_-_27462351 | 0.08 |
ENST00000448850.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr2_-_211179883 | 0.08 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr13_-_25746416 | 0.08 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr11_+_64323156 | 0.08 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr10_-_104597286 | 0.08 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr1_+_171227069 | 0.07 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr2_-_37193606 | 0.07 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chrY_-_20935572 | 0.07 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr12_-_10251603 | 0.07 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr20_-_1373682 | 0.07 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr7_-_56160625 | 0.07 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr11_+_107461948 | 0.07 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr10_+_103912137 | 0.07 |
ENST00000603742.1
ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr11_+_20044600 | 0.07 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr1_-_11036272 | 0.06 |
ENST00000520253.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr8_-_101117847 | 0.06 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chr16_-_18911366 | 0.06 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_+_90259593 | 0.06 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr19_-_7812446 | 0.06 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr4_+_71091786 | 0.06 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr16_+_33006369 | 0.06 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr14_-_107114267 | 0.06 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr14_+_101123580 | 0.06 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr19_-_50169064 | 0.06 |
ENST00000593337.1
ENST00000598808.1 ENST00000600453.1 ENST00000593818.1 ENST00000597198.1 ENST00000601809.1 ENST00000377139.3 |
IRF3
|
interferon regulatory factor 3 |
| chr19_-_50168962 | 0.06 |
ENST00000599223.1
ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3
|
interferon regulatory factor 3 |
| chr1_-_157522180 | 0.06 |
ENST00000356953.4
ENST00000368188.2 ENST00000368190.3 ENST00000368189.3 |
FCRL5
|
Fc receptor-like 5 |
| chrY_-_13524717 | 0.06 |
ENST00000331172.6
|
SLC9B1P1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr17_-_39646116 | 0.06 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr12_-_95044309 | 0.06 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr1_+_367640 | 0.06 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr4_+_56719782 | 0.05 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chrY_+_20708557 | 0.05 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr12_-_110883346 | 0.05 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_+_20044096 | 0.05 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr8_-_7220490 | 0.05 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr6_+_167704838 | 0.05 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr8_-_124749609 | 0.05 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr12_-_11287243 | 0.05 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr3_-_110612323 | 0.05 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr10_-_126694575 | 0.05 |
ENST00000334808.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr17_-_44657017 | 0.04 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr11_-_47870091 | 0.04 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr17_-_34257731 | 0.04 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr11_+_55594695 | 0.04 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2 |
| chr15_-_35088340 | 0.04 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr17_-_34257771 | 0.04 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr3_-_110612059 | 0.04 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr7_+_143657027 | 0.04 |
ENST00000392899.1
|
OR2F1
|
olfactory receptor, family 2, subfamily F, member 1 (gene/pseudogene) |
| chr12_-_10251576 | 0.04 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr2_-_29297127 | 0.04 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr8_-_13372253 | 0.04 |
ENST00000316609.5
|
DLC1
|
deleted in liver cancer 1 |
| chr4_-_152682129 | 0.04 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr4_-_145061788 | 0.04 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr6_+_167704798 | 0.04 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr1_-_71513471 | 0.04 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr2_-_106013400 | 0.04 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr19_-_40228657 | 0.04 |
ENST00000221804.4
|
CLC
|
Charcot-Leyden crystal galectin |
| chr3_-_38835501 | 0.04 |
ENST00000449082.2
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit |
| chr3_-_49158218 | 0.04 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr4_+_154073469 | 0.04 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr11_+_60609537 | 0.04 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr12_-_57873631 | 0.04 |
ENST00000393791.3
ENST00000356411.2 ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr13_-_103426112 | 0.04 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.2 | 1.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.5 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 2.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.3 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |