Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
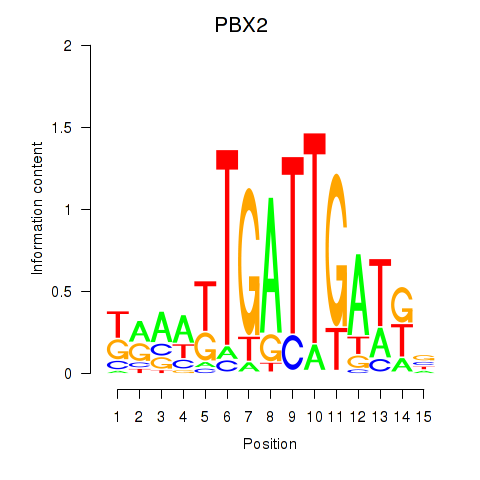
Results for PBX2
Z-value: 0.43
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg19_v2_chr6_-_32157947_32157992 | 0.81 | 8.1e-08 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_31440377 | 2.04 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr14_-_55658252 | 2.01 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr14_-_55658323 | 1.91 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr2_+_234104079 | 1.65 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr2_-_190044480 | 1.50 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr12_-_28124903 | 1.30 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr12_-_28125638 | 1.07 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr15_+_67418047 | 0.83 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr11_-_2162162 | 0.69 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr4_-_100065419 | 0.63 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr7_+_134528635 | 0.63 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr2_+_234601512 | 0.59 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr11_+_69455855 | 0.55 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr4_+_37962018 | 0.51 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr2_+_204732666 | 0.48 |
ENST00000295854.6
ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr11_+_62432777 | 0.47 |
ENST00000532971.1
|
METTL12
|
methyltransferase like 12 |
| chrX_+_46696372 | 0.46 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr2_+_33661382 | 0.41 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_-_29151794 | 0.37 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr1_+_207038699 | 0.33 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr2_+_20646824 | 0.32 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr1_+_101702417 | 0.32 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr2_-_99917639 | 0.31 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr9_-_128246769 | 0.30 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr5_+_54320078 | 0.30 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr2_+_204732487 | 0.29 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_+_188889737 | 0.28 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr14_+_27342334 | 0.24 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr14_+_64854958 | 0.22 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr1_+_62417957 | 0.20 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chrX_+_119737806 | 0.20 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr10_-_45474237 | 0.19 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr11_-_104769141 | 0.19 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr2_+_113033164 | 0.16 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr3_+_159943362 | 0.14 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr16_+_1128781 | 0.14 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr3_-_129158850 | 0.12 |
ENST00000503197.1
ENST00000249910.1 ENST00000429544.2 ENST00000507208.1 |
MBD4
|
methyl-CpG binding domain protein 4 |
| chr4_-_100065440 | 0.11 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr10_+_124320195 | 0.11 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr11_-_111794446 | 0.11 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_-_129158676 | 0.11 |
ENST00000393278.2
|
MBD4
|
methyl-CpG binding domain protein 4 |
| chr18_-_72920372 | 0.08 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr10_+_124320156 | 0.08 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_+_234602305 | 0.07 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_44396000 | 0.04 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr2_+_27255806 | 0.02 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr3_-_52864680 | 0.01 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.5 | 1.6 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.5 | 1.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.8 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 2.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.7 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.2 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 2.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 4.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |