Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
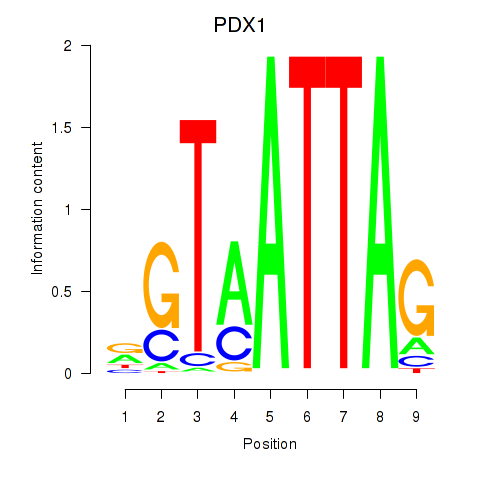
Results for PDX1
Z-value: 0.61
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | 0.31 | 1.0e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_152386732 | 1.25 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr12_+_107712173 | 0.81 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr7_-_44229022 | 0.73 |
ENST00000403799.3
|
GCK
|
glucokinase (hexokinase 4) |
| chr20_+_60174827 | 0.68 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr9_-_14722715 | 0.68 |
ENST00000380911.3
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr15_+_75080883 | 0.62 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr4_-_109541539 | 0.60 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr1_+_62439037 | 0.59 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr8_-_133123406 | 0.58 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr18_+_29027696 | 0.58 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr14_+_22985251 | 0.57 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr1_+_84609944 | 0.57 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_38911580 | 0.56 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr3_-_57233966 | 0.55 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr18_+_28898052 | 0.53 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr12_-_57328187 | 0.50 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr17_-_39150385 | 0.45 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr1_+_160160346 | 0.45 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr4_-_139163491 | 0.44 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr15_-_55562479 | 0.44 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_160160283 | 0.44 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_-_112127981 | 0.43 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr8_-_131399110 | 0.41 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr6_-_32157947 | 0.41 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr15_-_55563072 | 0.41 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_8285405 | 0.40 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr5_+_135394840 | 0.40 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr9_-_123639304 | 0.39 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr4_+_86525299 | 0.39 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_+_15699286 | 0.38 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr4_-_140544386 | 0.38 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr13_-_99667960 | 0.38 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr2_+_48541776 | 0.38 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr8_+_7783859 | 0.37 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
| chr12_-_54813229 | 0.37 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr11_+_65554493 | 0.37 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr9_+_125133315 | 0.36 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_+_56325812 | 0.36 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr17_-_18430160 | 0.36 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr10_-_48416849 | 0.35 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr9_+_135457530 | 0.35 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr11_-_8290263 | 0.35 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr4_-_69111401 | 0.35 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr5_+_126984710 | 0.35 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr17_-_9694614 | 0.35 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr12_-_6233828 | 0.35 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr8_-_42234745 | 0.34 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr9_-_136933615 | 0.34 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr14_-_57272366 | 0.33 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr12_-_86650077 | 0.33 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_-_77889860 | 0.33 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr12_-_91576429 | 0.32 |
ENST00000552145.1
ENST00000546745.1 |
DCN
|
decorin |
| chr6_+_34204642 | 0.32 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr20_+_42984330 | 0.32 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr2_-_208031943 | 0.31 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr9_-_123639600 | 0.31 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr7_+_129015484 | 0.30 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_-_100860851 | 0.30 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr3_-_98235962 | 0.30 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr9_+_125132803 | 0.30 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_-_55562582 | 0.30 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_+_28624878 | 0.29 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr3_-_143567262 | 0.29 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr12_-_53171128 | 0.29 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr11_-_82444892 | 0.28 |
ENST00000329203.3
|
FAM181B
|
family with sequence similarity 181, member B |
| chr4_+_70796784 | 0.28 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr9_-_123639445 | 0.27 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr21_+_17792672 | 0.27 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_+_30697298 | 0.27 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr15_+_58430567 | 0.26 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr10_+_119301928 | 0.26 |
ENST00000553456.3
|
EMX2
|
empty spiracles homeobox 2 |
| chr4_-_116034979 | 0.26 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr17_-_46690839 | 0.26 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chrX_-_20236970 | 0.26 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr15_+_58430368 | 0.25 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr7_+_50348268 | 0.25 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr20_+_62795827 | 0.25 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr17_-_27418537 | 0.25 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr7_-_14028488 | 0.24 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr12_+_28410128 | 0.24 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_21676442 | 0.24 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr9_-_28670283 | 0.23 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr14_-_36988882 | 0.23 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr18_+_59000815 | 0.22 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr7_-_81399411 | 0.22 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_45168875 | 0.22 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr4_-_39979576 | 0.22 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr7_-_81399355 | 0.22 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_-_123377880 | 0.22 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr7_-_27169801 | 0.21 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr20_+_43538692 | 0.21 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr8_-_7243080 | 0.21 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr7_-_14029283 | 0.21 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr18_-_30353025 | 0.21 |
ENST00000359358.4
|
KLHL14
|
kelch-like family member 14 |
| chr6_+_167704838 | 0.21 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr12_-_52967600 | 0.20 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr17_-_57229155 | 0.20 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr15_+_63188009 | 0.20 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr12_+_81110684 | 0.20 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr20_-_31124186 | 0.20 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chrX_+_7137475 | 0.20 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr4_+_141445311 | 0.20 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr21_-_35899113 | 0.20 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr14_+_100485712 | 0.20 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr20_+_45947246 | 0.19 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr2_-_112237835 | 0.19 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr2_+_196313239 | 0.19 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr6_+_29079668 | 0.19 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr2_+_90060377 | 0.19 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr20_+_43538756 | 0.19 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr17_-_39156138 | 0.19 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr15_+_62853562 | 0.18 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr2_-_99871570 | 0.18 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr3_-_74570291 | 0.18 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr10_+_63808970 | 0.18 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr13_+_102104952 | 0.18 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_+_167704798 | 0.18 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr8_+_77593448 | 0.18 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr19_-_7968427 | 0.18 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr11_-_115630900 | 0.18 |
ENST00000537070.1
ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900
|
long intergenic non-protein coding RNA 900 |
| chr3_-_39321512 | 0.18 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr2_+_87769459 | 0.17 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr10_-_21435488 | 0.17 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr3_+_111718173 | 0.17 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr14_+_22554680 | 0.17 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr4_-_103749179 | 0.17 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_9953214 | 0.17 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr2_-_89459813 | 0.16 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr3_-_108248169 | 0.16 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr17_+_74846230 | 0.16 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr21_+_17442799 | 0.16 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr22_-_32860427 | 0.16 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chr2_-_227050079 | 0.16 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr8_+_77593474 | 0.16 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr1_+_28586006 | 0.16 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr8_+_9953061 | 0.15 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr4_-_103749205 | 0.15 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_37457350 | 0.15 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr1_+_1846519 | 0.15 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chrX_-_110655306 | 0.15 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr1_-_151965048 | 0.15 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr4_-_119759795 | 0.14 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr1_-_190446759 | 0.14 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_66536196 | 0.14 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr10_+_24738355 | 0.14 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr19_-_47137942 | 0.14 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr10_+_11047259 | 0.14 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr4_+_71587669 | 0.14 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_-_100912785 | 0.13 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr7_-_73038822 | 0.13 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr8_+_22424551 | 0.13 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr14_+_61654271 | 0.13 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr1_-_68698222 | 0.13 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr7_-_73038867 | 0.13 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr15_+_93443419 | 0.13 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_+_69985792 | 0.13 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_12834984 | 0.12 |
ENST00000357726.4
|
PRAMEF12
|
PRAME family member 12 |
| chr11_-_115375107 | 0.12 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr14_-_69261310 | 0.12 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr4_-_66536057 | 0.11 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr1_-_13002348 | 0.11 |
ENST00000355096.2
|
PRAMEF6
|
PRAME family member 6 |
| chr4_-_103749313 | 0.11 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_+_641178 | 0.11 |
ENST00000166133.3
|
FGF22
|
fibroblast growth factor 22 |
| chr21_+_17443521 | 0.11 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_6533729 | 0.11 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_95236551 | 0.11 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr6_-_111136513 | 0.11 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr21_+_17443434 | 0.10 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_+_69854027 | 0.10 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_+_169013666 | 0.10 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chrX_-_18690210 | 0.10 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr7_-_81399438 | 0.10 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_+_136501478 | 0.10 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr18_+_3447572 | 0.10 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_-_9630463 | 0.10 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr3_+_136649311 | 0.10 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr1_-_68698197 | 0.10 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr12_+_81101277 | 0.10 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chrX_-_64196376 | 0.10 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr19_+_13049413 | 0.09 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr19_+_11071546 | 0.09 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr8_+_92261516 | 0.09 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr15_+_67841330 | 0.09 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chrX_-_64196307 | 0.09 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr16_-_52640834 | 0.09 |
ENST00000510238.3
|
CASC16
|
cancer susceptibility candidate 16 (non-protein coding) |
| chr4_-_103749105 | 0.09 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_234667504 | 0.09 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr5_-_142780280 | 0.09 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_102312517 | 0.09 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr15_+_80351910 | 0.09 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr14_-_47351391 | 0.09 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr6_-_29399744 | 0.09 |
ENST00000377154.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chrX_-_100129128 | 0.09 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr12_-_56106060 | 0.09 |
ENST00000452168.2
|
ITGA7
|
integrin, alpha 7 |
| chr12_-_56321649 | 0.09 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr7_-_99717463 | 0.08 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrX_-_64196351 | 0.08 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chrX_+_133930798 | 0.08 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr1_+_198608146 | 0.08 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr12_-_32908809 | 0.08 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.7 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.4 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.7 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.2 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.2 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0051794 | catagen(GO:0042637) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.3 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |