Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PGR
Z-value: 0.82
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_101000445_101000465, hg19_v2_chr11_-_100999775_100999801 | -0.10 | 6.1e-01 | Click! |
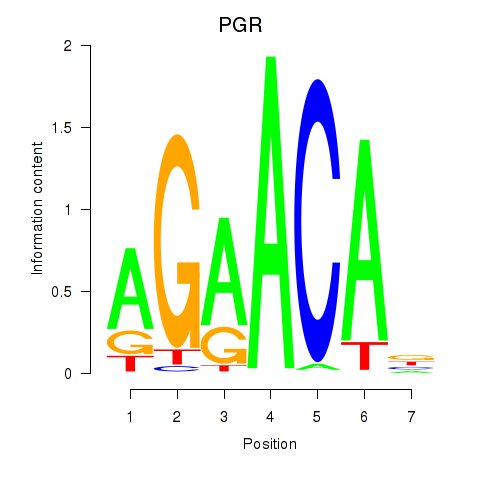
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_27062860 | 2.87 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_-_71551652 | 2.61 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr7_-_5821225 | 2.60 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr15_-_49255632 | 2.40 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr19_-_9003586 | 1.88 |
ENST00000380951.5
|
MUC16
|
mucin 16, cell surface associated |
| chr5_-_160279207 | 1.67 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr5_+_140261703 | 1.58 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr6_-_46922659 | 1.55 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr5_-_121413974 | 1.41 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr8_+_40010989 | 1.31 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr7_+_73442487 | 1.26 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr11_+_111169565 | 1.16 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr11_+_6897856 | 1.12 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr19_-_40440533 | 1.10 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr7_+_73442422 | 1.09 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr16_+_78056412 | 1.07 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr12_-_91505608 | 1.02 |
ENST00000266718.4
|
LUM
|
lumican |
| chr3_-_151102529 | 0.93 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr6_-_32908792 | 0.92 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_-_145039835 | 0.88 |
ENST00000533259.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr7_+_73442457 | 0.83 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chrX_+_16964985 | 0.80 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr3_-_114343768 | 0.79 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_156828810 | 0.77 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr7_+_30960915 | 0.76 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr5_+_102201509 | 0.75 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr19_+_13135731 | 0.74 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_+_191334212 | 0.72 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr4_-_154710210 | 0.67 |
ENST00000274063.4
|
SFRP2
|
secreted frizzled-related protein 2 |
| chr4_+_86525299 | 0.67 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_+_131438443 | 0.66 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr5_+_67584174 | 0.66 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr16_-_3350614 | 0.66 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr10_-_48416849 | 0.65 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr10_-_28571015 | 0.64 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_+_73442102 | 0.62 |
ENST00000445912.1
ENST00000252034.7 |
ELN
|
elastin |
| chr5_-_124081008 | 0.61 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr5_+_102201430 | 0.60 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_95975672 | 0.59 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr14_+_24674926 | 0.59 |
ENST00000339917.5
ENST00000556621.1 ENST00000287913.6 ENST00000428351.2 ENST00000555092.1 |
TSSK4
|
testis-specific serine kinase 4 |
| chr6_+_39760129 | 0.59 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_+_56473628 | 0.58 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr3_-_58563094 | 0.58 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr19_+_13135386 | 0.58 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_-_154831779 | 0.58 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr9_+_127615733 | 0.58 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr19_+_13135790 | 0.56 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_+_105603683 | 0.56 |
ENST00000317716.9
|
CDHR3
|
cadherin-related family member 3 |
| chr12_-_88974236 | 0.55 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr1_-_145039949 | 0.54 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr3_-_114035026 | 0.53 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr19_+_41119323 | 0.52 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr20_+_52105495 | 0.52 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr12_-_81763127 | 0.52 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_-_81763184 | 0.51 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_+_6502675 | 0.51 |
ENST00000254616.6
ENST00000530751.1 |
TIMM10B
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr6_-_32908765 | 0.51 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr2_-_175711133 | 0.49 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr4_-_88450612 | 0.49 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr5_+_102201722 | 0.48 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_149051194 | 0.47 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr20_+_23016057 | 0.47 |
ENST00000255008.3
|
SSTR4
|
somatostatin receptor 4 |
| chr6_+_30951487 | 0.47 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr11_+_123396528 | 0.46 |
ENST00000322282.7
ENST00000529750.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr10_+_104178946 | 0.45 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr17_-_71088797 | 0.45 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr7_-_92855762 | 0.45 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr5_+_102201687 | 0.45 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_38177575 | 0.44 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr21_-_34185944 | 0.44 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr17_+_73997796 | 0.44 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr20_+_138089 | 0.43 |
ENST00000382388.3
|
DEFB127
|
defensin, beta 127 |
| chr19_+_16771936 | 0.43 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr5_-_94417339 | 0.43 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_27107053 | 0.42 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr15_-_43877062 | 0.41 |
ENST00000381885.1
ENST00000396923.3 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr3_-_112218378 | 0.41 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr19_-_42947121 | 0.41 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chrX_-_112084043 | 0.41 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr17_+_7761013 | 0.40 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr15_+_31658349 | 0.40 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr3_-_114343039 | 0.40 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_22089845 | 0.40 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_-_67955426 | 0.39 |
ENST00000554480.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_47010061 | 0.39 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr3_+_149530836 | 0.39 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr1_-_167906277 | 0.39 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr1_-_85870177 | 0.38 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr1_+_154975258 | 0.38 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr2_-_158182410 | 0.37 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr22_-_31688381 | 0.37 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_+_116225999 | 0.37 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_-_191000172 | 0.37 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr2_-_216300784 | 0.36 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr12_+_58003935 | 0.36 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr19_-_51529849 | 0.35 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr7_+_105603657 | 0.35 |
ENST00000542731.1
ENST00000343407.5 |
CDHR3
|
cadherin-related family member 3 |
| chr22_-_31688431 | 0.35 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr17_-_56621665 | 0.34 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr11_-_73693875 | 0.34 |
ENST00000536983.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr7_-_150946015 | 0.34 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chrX_+_130192216 | 0.33 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr10_-_104178857 | 0.32 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr2_+_219537015 | 0.32 |
ENST00000440309.1
ENST00000424080.1 |
STK36
|
serine/threonine kinase 36 |
| chr5_-_38557561 | 0.31 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr6_-_132939317 | 0.31 |
ENST00000275191.2
|
TAAR2
|
trace amine associated receptor 2 |
| chr3_-_124839648 | 0.31 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr15_-_45406385 | 0.30 |
ENST00000389039.6
|
DUOX2
|
dual oxidase 2 |
| chrX_+_69672136 | 0.30 |
ENST00000374355.3
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr16_+_202686 | 0.30 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr6_+_30131318 | 0.30 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr9_-_140196703 | 0.30 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr7_+_138145145 | 0.29 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr11_-_124806297 | 0.29 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr9_+_2159850 | 0.29 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_-_49482907 | 0.29 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr15_+_31619013 | 0.29 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr1_-_145039771 | 0.29 |
ENST00000493130.2
ENST00000532801.1 ENST00000478649.2 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr5_-_146435501 | 0.28 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr22_+_31644388 | 0.28 |
ENST00000333611.4
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr13_+_102104952 | 0.28 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr16_-_66952779 | 0.28 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_-_76782371 | 0.28 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr9_-_34665983 | 0.28 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr18_+_6729698 | 0.28 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr5_-_146435572 | 0.28 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr20_+_34129770 | 0.28 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chrX_-_100641155 | 0.27 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr6_-_159466042 | 0.27 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr17_-_4463856 | 0.27 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr9_+_1051481 | 0.27 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr17_+_46970134 | 0.27 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr16_-_66952742 | 0.26 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr4_-_105416039 | 0.26 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr5_+_156607829 | 0.26 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr17_-_26697304 | 0.26 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr14_-_107219365 | 0.25 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr11_-_8832182 | 0.25 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr9_+_82188077 | 0.25 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_+_22985251 | 0.25 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr3_+_29323043 | 0.25 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr16_-_3306587 | 0.25 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr3_+_25469802 | 0.25 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr1_+_154975110 | 0.25 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr2_+_219536749 | 0.24 |
ENST00000295709.3
ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36
|
serine/threonine kinase 36 |
| chr5_-_146435694 | 0.24 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr20_+_59654146 | 0.24 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr3_+_137906109 | 0.24 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr1_-_26394114 | 0.24 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr5_-_76935513 | 0.24 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr4_-_87028478 | 0.23 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_19144243 | 0.23 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr19_+_10531150 | 0.23 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr8_+_10530155 | 0.23 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr1_-_166944561 | 0.23 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr7_+_120590803 | 0.23 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr20_+_43514315 | 0.23 |
ENST00000353703.4
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chr5_+_137419581 | 0.22 |
ENST00000506684.1
ENST00000504809.1 ENST00000398754.1 |
WNT8A
|
wingless-type MMTV integration site family, member 8A |
| chr4_+_146539415 | 0.22 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr1_+_41174988 | 0.22 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_+_46970178 | 0.22 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_+_50146747 | 0.22 |
ENST00000547798.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_-_48136217 | 0.22 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr5_+_139505520 | 0.22 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr7_+_22766766 | 0.22 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr20_+_43514320 | 0.22 |
ENST00000372839.3
ENST00000428262.1 ENST00000445830.1 |
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chrX_-_32173579 | 0.22 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr17_+_59477233 | 0.22 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr9_+_131683174 | 0.22 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr17_-_40540586 | 0.21 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr13_+_73632897 | 0.21 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr15_+_65843130 | 0.21 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr6_-_89927151 | 0.21 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr12_+_56661461 | 0.21 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr17_+_46970127 | 0.21 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_-_6484715 | 0.21 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr5_+_138629417 | 0.21 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chrX_+_134887233 | 0.20 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr2_-_89266286 | 0.20 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr11_-_6502534 | 0.20 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr3_+_25469724 | 0.20 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr20_+_23420322 | 0.20 |
ENST00000347397.1
|
CSTL1
|
cystatin-like 1 |
| chr12_+_57849048 | 0.19 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr5_+_133842243 | 0.19 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr17_-_15522826 | 0.19 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr20_+_23420885 | 0.19 |
ENST00000246020.2
|
CSTL1
|
cystatin-like 1 |
| chr5_+_72469014 | 0.19 |
ENST00000296776.5
|
TMEM174
|
transmembrane protein 174 |
| chrX_-_131262048 | 0.19 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chr1_+_161736072 | 0.19 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr10_+_63808970 | 0.18 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr5_+_138629337 | 0.18 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr13_+_102104980 | 0.18 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr9_+_125281420 | 0.18 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr1_-_32210275 | 0.18 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr15_-_42783303 | 0.18 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr1_+_152691998 | 0.18 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr1_+_16370271 | 0.18 |
ENST00000375679.4
|
CLCNKB
|
chloride channel, voltage-sensitive Kb |
| chr11_-_6502580 | 0.18 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr1_-_179112189 | 0.18 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_+_36923830 | 0.17 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.5 | 1.6 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.3 | 2.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.8 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.2 | 0.7 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 1.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 0.6 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.2 | 0.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.9 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.4 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.3 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.4 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.5 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 2.7 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.1 | 1.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.2 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 2.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.7 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 3.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.4 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 2.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0061042 | heparan sulfate proteoglycan catabolic process(GO:0030200) vascular wound healing(GO:0061042) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.0 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.6 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 0.8 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 0.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 3.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.0 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 2.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 1.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 0.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 0.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.1 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |