Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PKNOX1_TGIF2
Z-value: 0.51
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
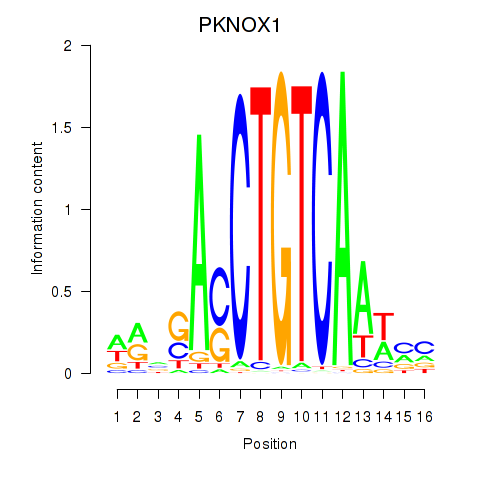
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
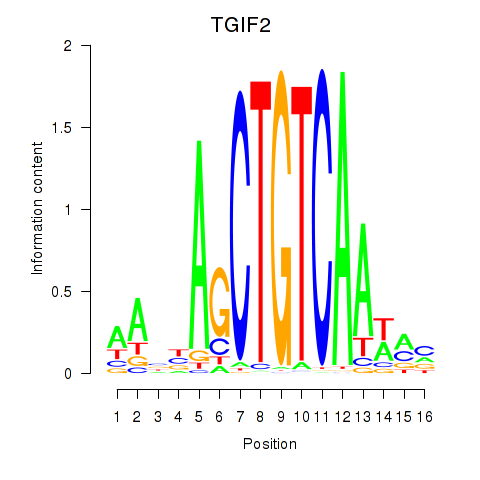
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2 | hg19_v2_chr20_+_35201857_35201891, hg19_v2_chr20_+_35201993_35202050 | 0.33 | 7.2e-02 | Click! |
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | 0.19 | 3.2e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_34185944 | 1.36 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_+_827553 | 1.08 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr17_+_68071458 | 1.04 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr13_-_39564993 | 1.04 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr17_+_68071389 | 1.03 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr7_-_99869799 | 0.94 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr6_+_163148973 | 0.86 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr18_+_3449330 | 0.81 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_74779744 | 0.73 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr21_-_34185989 | 0.63 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr6_-_24911195 | 0.59 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr2_-_74780176 | 0.59 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr4_-_105416039 | 0.52 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr22_+_39052632 | 0.52 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr5_-_132073111 | 0.52 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr15_-_37391614 | 0.47 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr15_-_37392086 | 0.46 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr17_+_39405939 | 0.43 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr12_+_55248289 | 0.42 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr14_+_100842735 | 0.41 |
ENST00000554998.1
ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25
|
WD repeat domain 25 |
| chr4_-_47840112 | 0.41 |
ENST00000504584.1
|
CORIN
|
corin, serine peptidase |
| chr4_-_83483395 | 0.41 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr1_+_63989004 | 0.40 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr19_+_49496705 | 0.39 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr21_-_43735446 | 0.36 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr1_+_207262170 | 0.36 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr21_-_34186006 | 0.36 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr1_-_22469459 | 0.36 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr16_+_618837 | 0.35 |
ENST00000409439.2
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_+_207262578 | 0.35 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr1_+_207262627 | 0.35 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr18_+_905104 | 0.34 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr1_+_207262540 | 0.34 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr19_-_10764509 | 0.34 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr16_+_2588012 | 0.32 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr12_-_31882108 | 0.32 |
ENST00000281471.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr18_+_54318566 | 0.32 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr14_-_92198403 | 0.31 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr3_-_182880541 | 0.31 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr1_-_53163992 | 0.31 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr3_-_113160334 | 0.31 |
ENST00000393845.2
ENST00000295868.2 |
WDR52
|
WD repeat domain 52 |
| chr3_+_52828805 | 0.29 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr18_-_54305658 | 0.29 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr4_-_46391805 | 0.29 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr12_-_91398796 | 0.29 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr17_+_76374714 | 0.29 |
ENST00000262764.6
ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1
|
phosphatidylglycerophosphate synthase 1 |
| chrX_+_49019061 | 0.29 |
ENST00000376339.1
ENST00000425661.2 ENST00000458388.1 ENST00000412696.2 |
MAGIX
|
MAGI family member, X-linked |
| chr4_-_149365827 | 0.28 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr22_-_37172111 | 0.28 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr16_+_4784273 | 0.27 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr22_-_37172191 | 0.27 |
ENST00000340630.5
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr4_+_86396265 | 0.27 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_+_210406121 | 0.26 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr3_-_183273477 | 0.26 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr19_+_49496782 | 0.26 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_+_2587998 | 0.26 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr14_-_94856987 | 0.26 |
ENST00000449399.3
ENST00000404814.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_+_3668962 | 0.25 |
ENST00000294600.2
|
CCDC27
|
coiled-coil domain containing 27 |
| chr16_-_50715196 | 0.25 |
ENST00000423026.2
|
SNX20
|
sorting nexin 20 |
| chr2_+_234959376 | 0.25 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr16_+_30212378 | 0.24 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr12_-_67197760 | 0.23 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr6_+_26501449 | 0.23 |
ENST00000244513.6
|
BTN1A1
|
butyrophilin, subfamily 1, member A1 |
| chr16_+_16484691 | 0.23 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr9_-_86432547 | 0.23 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr9_-_124989804 | 0.22 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr17_-_26220366 | 0.22 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr7_+_23637763 | 0.22 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chrX_-_101397433 | 0.22 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chr19_+_49497121 | 0.22 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr5_+_125758813 | 0.22 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr7_+_73442487 | 0.21 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr6_-_132967142 | 0.21 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr6_-_31651817 | 0.21 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr12_-_121734489 | 0.20 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr14_-_50474238 | 0.20 |
ENST00000399206.1
|
C14orf182
|
chromosome 14 open reading frame 182 |
| chr16_-_69418649 | 0.20 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr5_+_125758865 | 0.20 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr14_-_94857004 | 0.19 |
ENST00000557492.1
ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr12_-_104531785 | 0.19 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr11_+_27062502 | 0.19 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr16_-_30457048 | 0.19 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr11_-_5255861 | 0.18 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr14_-_94856951 | 0.18 |
ENST00000553327.1
ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr6_-_32920794 | 0.18 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr16_+_2587965 | 0.18 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr4_+_123653807 | 0.17 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_+_246729724 | 0.17 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr7_+_73442422 | 0.17 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr17_+_38296576 | 0.17 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr4_+_146539415 | 0.16 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr11_+_27062272 | 0.16 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_104458235 | 0.15 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr3_-_3152031 | 0.15 |
ENST00000383846.1
ENST00000427088.1 ENST00000446632.2 ENST00000438560.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr1_-_54872059 | 0.15 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr7_-_156685890 | 0.15 |
ENST00000353442.5
|
LMBR1
|
limb development membrane protein 1 |
| chr12_-_121019165 | 0.15 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr22_+_17082732 | 0.15 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr12_+_122667658 | 0.15 |
ENST00000339777.4
ENST00000425921.1 |
LRRC43
|
leucine rich repeat containing 43 |
| chr1_+_145549203 | 0.14 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr3_-_27763803 | 0.14 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr3_-_3221358 | 0.13 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr19_+_11909329 | 0.13 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr2_+_16080659 | 0.13 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr18_+_39535152 | 0.13 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr16_-_1429627 | 0.13 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr1_+_154966058 | 0.13 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr7_+_39605966 | 0.13 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr9_-_115653176 | 0.13 |
ENST00000374228.4
|
SLC46A2
|
solute carrier family 46, member 2 |
| chr10_+_49514698 | 0.13 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr7_+_73442457 | 0.13 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr1_+_171217622 | 0.13 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr11_+_112046190 | 0.13 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr17_+_39382900 | 0.13 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr16_-_50715239 | 0.12 |
ENST00000330943.4
ENST00000300590.3 |
SNX20
|
sorting nexin 20 |
| chr21_-_30257669 | 0.12 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr16_+_20817839 | 0.12 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr16_-_4784128 | 0.12 |
ENST00000592711.1
ENST00000590147.1 ENST00000304283.4 ENST00000592190.1 ENST00000589065.1 ENST00000585773.1 ENST00000450067.2 ENST00000592698.1 ENST00000586166.1 ENST00000586605.1 ENST00000592421.1 |
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr10_+_28966271 | 0.12 |
ENST00000375533.3
|
BAMBI
|
BMP and activin membrane-bound inhibitor |
| chr22_-_50219548 | 0.12 |
ENST00000404034.1
|
BRD1
|
bromodomain containing 1 |
| chr18_+_32073253 | 0.12 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr3_-_52488048 | 0.12 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr19_+_39759154 | 0.12 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr8_-_128960591 | 0.12 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr15_-_31283798 | 0.12 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr5_-_58882219 | 0.11 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_173043591 | 0.11 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr15_+_25068773 | 0.11 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr15_+_88120158 | 0.11 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr20_+_54987305 | 0.11 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr21_+_31768348 | 0.11 |
ENST00000355459.2
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr11_-_6440283 | 0.11 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_-_169587621 | 0.11 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr10_+_94608245 | 0.11 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr6_+_26124373 | 0.11 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr13_+_33160553 | 0.11 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr16_+_20817761 | 0.11 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr12_+_59989918 | 0.11 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr15_-_78538025 | 0.11 |
ENST00000560817.1
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_71454185 | 0.10 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr4_+_86396321 | 0.10 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_27764190 | 0.10 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_+_168841817 | 0.10 |
ENST00000356284.2
ENST00000354536.5 |
SMOC2
|
SPARC related modular calcium binding 2 |
| chr3_+_52719936 | 0.10 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_-_3151664 | 0.10 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr1_-_63988846 | 0.10 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr6_-_119031228 | 0.10 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr6_+_31865552 | 0.10 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr19_+_1000418 | 0.10 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chrX_-_39923656 | 0.10 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr4_+_48485341 | 0.10 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr9_-_134615443 | 0.10 |
ENST00000372195.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr16_+_4896659 | 0.10 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr20_+_54987168 | 0.10 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_-_246729544 | 0.09 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr4_+_88928777 | 0.09 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr2_+_113033164 | 0.09 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr5_+_43603229 | 0.09 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr3_+_185080908 | 0.09 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr11_-_101000445 | 0.09 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr2_-_203103281 | 0.09 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr7_+_63774321 | 0.09 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr7_+_99971068 | 0.09 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr6_+_31674639 | 0.09 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr19_-_45826125 | 0.09 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr19_-_12595586 | 0.08 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr5_-_169725231 | 0.08 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr3_+_150804676 | 0.08 |
ENST00000474524.1
ENST00000273432.4 |
MED12L
|
mediator complex subunit 12-like |
| chr19_-_39322299 | 0.08 |
ENST00000601094.1
ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr7_+_23637118 | 0.08 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr6_-_28220002 | 0.08 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr17_+_8191815 | 0.08 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr12_+_129028500 | 0.08 |
ENST00000315208.8
|
TMEM132C
|
transmembrane protein 132C |
| chr15_-_43785303 | 0.08 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_-_179112189 | 0.08 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr1_+_203765437 | 0.08 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr14_-_106330458 | 0.08 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr19_+_49375649 | 0.08 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr12_-_31479107 | 0.08 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr21_-_34144157 | 0.07 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr12_-_75784669 | 0.07 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr6_-_32140886 | 0.07 |
ENST00000395496.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr11_-_84028180 | 0.07 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_-_51425902 | 0.07 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr11_-_64052111 | 0.07 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr2_-_3504587 | 0.07 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr6_-_52926539 | 0.07 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr5_-_81046904 | 0.07 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr16_-_89556942 | 0.07 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr12_-_15114191 | 0.06 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_+_52800410 | 0.06 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr7_+_38217818 | 0.06 |
ENST00000009041.7
ENST00000544203.1 ENST00000434197.1 |
STARD3NL
|
STARD3 N-terminal like |
| chr15_+_57668695 | 0.06 |
ENST00000281282.5
|
CGNL1
|
cingulin-like 1 |
| chr17_-_34195889 | 0.06 |
ENST00000311880.2
|
C17orf66
|
chromosome 17 open reading frame 66 |
| chr3_+_111260954 | 0.06 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr2_-_179914760 | 0.06 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr17_-_34195862 | 0.06 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr11_+_64059464 | 0.06 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr14_-_69619689 | 0.06 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr13_-_46716969 | 0.06 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 0.9 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 1.3 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.4 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.4 | GO:0061054 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) renal vesicle induction(GO:0072034) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.3 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.2 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.6 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 0.8 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.9 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |