Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
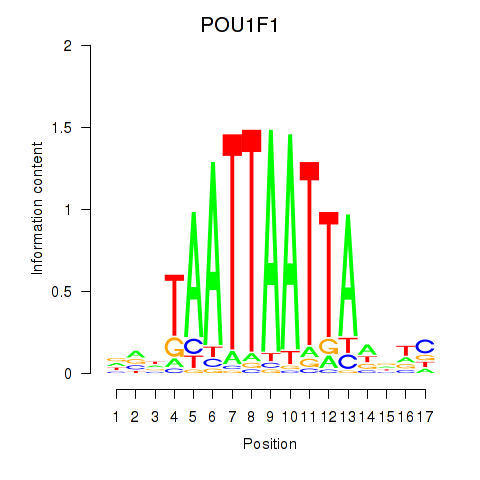
Results for POU1F1
Z-value: 0.53
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325612_87325654 | -0.30 | 1.1e-01 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13141097 | 1.96 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr16_-_30122717 | 1.61 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr5_+_147691979 | 1.01 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr1_+_115572415 | 0.82 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr17_-_5321549 | 0.77 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr10_+_94352956 | 0.64 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr8_-_91095099 | 0.62 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr1_+_192127578 | 0.56 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr9_+_125133315 | 0.52 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_-_43969796 | 0.47 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr18_-_59274139 | 0.47 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr10_+_52152766 | 0.46 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr18_+_29027696 | 0.45 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr9_+_18474204 | 0.44 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS-like 1 |
| chrX_-_11308598 | 0.44 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr4_-_153601136 | 0.40 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr4_-_159956333 | 0.40 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr19_-_36001113 | 0.40 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr2_-_190044480 | 0.40 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr12_+_131438443 | 0.39 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr15_+_67418047 | 0.38 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr1_-_205391178 | 0.38 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr19_-_58864848 | 0.34 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_35201826 | 0.34 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_52761262 | 0.34 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chrX_-_55057403 | 0.33 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr3_-_48130707 | 0.32 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr14_-_80697396 | 0.32 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr8_-_110986918 | 0.31 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr17_-_64225508 | 0.31 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_+_87012753 | 0.30 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr15_+_69365265 | 0.30 |
ENST00000415504.1
|
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr9_+_125132803 | 0.29 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_98900132 | 0.28 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_-_87342564 | 0.26 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr20_+_3776371 | 0.26 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr12_+_93096759 | 0.25 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr13_+_32313658 | 0.25 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr6_-_134861089 | 0.25 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr4_+_71296204 | 0.25 |
ENST00000413702.1
|
MUC7
|
mucin 7, secreted |
| chr6_+_26087646 | 0.25 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chrX_+_84258832 | 0.24 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr8_+_11961898 | 0.24 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr11_-_5323226 | 0.24 |
ENST00000380224.1
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4 |
| chr12_+_12510352 | 0.24 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr18_+_21269556 | 0.23 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr5_-_88119580 | 0.23 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr5_+_81575281 | 0.23 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr9_-_34729457 | 0.23 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chr6_-_49712123 | 0.23 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr10_+_62538089 | 0.22 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr9_+_111624577 | 0.22 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr2_-_43266680 | 0.22 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr9_-_21351377 | 0.22 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr14_+_31046959 | 0.22 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr18_+_21269404 | 0.22 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr7_+_107224364 | 0.22 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr12_+_93096619 | 0.22 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr4_+_74347400 | 0.22 |
ENST00000226355.3
|
AFM
|
afamin |
| chr4_+_76481258 | 0.21 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr9_+_12693336 | 0.21 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr2_-_163008903 | 0.21 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr10_-_75226166 | 0.21 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr14_-_101295407 | 0.21 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr7_+_129932974 | 0.20 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chrX_-_32173579 | 0.20 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr15_+_58702742 | 0.20 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr11_-_117186946 | 0.20 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr6_+_26087509 | 0.20 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr12_-_11287243 | 0.20 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr2_-_231860596 | 0.20 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr3_+_130650738 | 0.20 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chrX_+_154113317 | 0.19 |
ENST00000354461.2
|
H2AFB1
|
H2A histone family, member B1 |
| chr7_-_14026063 | 0.19 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr4_+_96012614 | 0.18 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_+_58655461 | 0.18 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chrX_+_153533275 | 0.18 |
ENST00000426989.1
ENST00000426203.1 ENST00000369912.2 |
TKTL1
|
transketolase-like 1 |
| chr10_-_99052382 | 0.18 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr19_-_56826157 | 0.18 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr20_+_30102231 | 0.18 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chrX_-_71458802 | 0.17 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr15_-_62937380 | 0.17 |
ENST00000560347.1
ENST00000558940.1 |
RP11-625H11.1
|
Uncharacterized protein |
| chr16_-_67217844 | 0.17 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr12_-_7596735 | 0.17 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr6_-_49712147 | 0.17 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_+_25205666 | 0.17 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr1_+_8378140 | 0.16 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr18_+_616672 | 0.16 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr12_-_7656357 | 0.16 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr17_+_61086917 | 0.16 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_-_154689596 | 0.16 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr6_+_47749718 | 0.16 |
ENST00000489301.2
ENST00000371211.2 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr11_+_55650773 | 0.16 |
ENST00000449290.2
|
TRIM51
|
tripartite motif-containing 51 |
| chr1_-_67266939 | 0.15 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr11_-_107729887 | 0.15 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr6_-_49604545 | 0.15 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr5_-_36301984 | 0.15 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr3_+_28390637 | 0.14 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr19_+_54466179 | 0.14 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr12_+_69753448 | 0.14 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr5_+_119867159 | 0.14 |
ENST00000505123.1
|
PRR16
|
proline rich 16 |
| chrX_+_154610428 | 0.14 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chr12_-_95510743 | 0.14 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chrX_-_77225135 | 0.14 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr12_-_86650045 | 0.13 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_17981462 | 0.13 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr8_-_57233103 | 0.13 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr4_+_74301880 | 0.13 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr5_-_93077293 | 0.13 |
ENST00000510627.4
|
POU5F2
|
POU domain class 5, transcription factor 2 |
| chr12_-_112123524 | 0.13 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr7_-_14026123 | 0.13 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr2_+_234826016 | 0.13 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_-_166930131 | 0.13 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr12_-_118797475 | 0.13 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr19_+_9296279 | 0.12 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr14_+_61654271 | 0.12 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr1_+_81771806 | 0.12 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr2_+_161993465 | 0.12 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_160823040 | 0.12 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr7_-_86688990 | 0.12 |
ENST00000450689.2
|
KIAA1324L
|
KIAA1324-like |
| chr1_+_149754227 | 0.12 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr1_-_197036364 | 0.11 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr11_+_55578854 | 0.11 |
ENST00000333973.2
|
OR5L1
|
olfactory receptor, family 5, subfamily L, member 1 |
| chr12_+_82347498 | 0.11 |
ENST00000550506.1
|
RP11-362A1.1
|
RP11-362A1.1 |
| chr9_-_21482312 | 0.11 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr6_+_114178512 | 0.11 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr5_+_60933634 | 0.11 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr19_+_16940198 | 0.11 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr3_-_160823158 | 0.11 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr15_+_64680003 | 0.11 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_-_45795145 | 0.10 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr10_-_28623368 | 0.10 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr19_+_52873166 | 0.10 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr7_+_99425633 | 0.10 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr3_+_160939050 | 0.10 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr2_+_152214098 | 0.10 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr17_+_45728427 | 0.10 |
ENST00000540627.1
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr4_+_186990298 | 0.10 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr5_+_115177178 | 0.10 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr4_+_3344141 | 0.10 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr10_+_18549645 | 0.10 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_-_53608289 | 0.10 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr1_+_109256067 | 0.09 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr17_+_1944790 | 0.09 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chrX_+_150565653 | 0.09 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr7_-_140482926 | 0.09 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr8_+_7801144 | 0.09 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr19_+_572543 | 0.09 |
ENST00000333511.3
ENST00000573216.1 ENST00000353555.4 |
BSG
|
basigin (Ok blood group) |
| chr19_-_22193731 | 0.09 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr16_+_28565230 | 0.09 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr6_-_109702885 | 0.08 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr17_-_41050716 | 0.08 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr20_+_55904815 | 0.08 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr5_-_111754948 | 0.08 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_-_191000172 | 0.08 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr4_-_185275104 | 0.08 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr8_+_70404996 | 0.08 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr12_-_15038779 | 0.08 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr11_-_89224488 | 0.08 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr4_-_139051839 | 0.08 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr6_+_154360476 | 0.08 |
ENST00000428397.2
|
OPRM1
|
opioid receptor, mu 1 |
| chr8_+_36641842 | 0.08 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr10_+_122610687 | 0.08 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr2_+_161993412 | 0.08 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_141957847 | 0.08 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr4_+_190861993 | 0.08 |
ENST00000524583.1
ENST00000531991.2 |
FRG1
|
FSHD region gene 1 |
| chr19_-_4535233 | 0.08 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr20_+_30697298 | 0.07 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr19_+_19144384 | 0.07 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr7_+_130126165 | 0.07 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr16_+_48278178 | 0.07 |
ENST00000285737.4
ENST00000535754.1 |
LONP2
|
lon peptidase 2, peroxisomal |
| chr6_+_29429217 | 0.07 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chrX_+_43515467 | 0.07 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr7_+_130126012 | 0.07 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr9_-_21239978 | 0.07 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr12_-_67197760 | 0.07 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr7_-_76829125 | 0.07 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr10_-_28571015 | 0.07 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_86650077 | 0.07 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_+_26104104 | 0.07 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr11_+_73498898 | 0.06 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr18_-_64271363 | 0.06 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr9_-_95056010 | 0.06 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr12_-_10978957 | 0.06 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr10_-_115904361 | 0.06 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_+_5526883 | 0.06 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chrX_-_106243451 | 0.06 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr22_-_32767017 | 0.06 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr7_-_112758665 | 0.06 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr9_-_95055956 | 0.06 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr9_-_85882145 | 0.06 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr12_+_8309630 | 0.06 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr15_-_22448819 | 0.06 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr1_-_247921982 | 0.06 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr6_-_136788001 | 0.06 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr1_-_211307404 | 0.06 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr12_+_113796347 | 0.06 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr6_-_132939317 | 0.05 |
ENST00000275191.2
|
TAAR2
|
trace amine associated receptor 2 |
| chr4_+_48833234 | 0.05 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr10_-_27529486 | 0.05 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.0 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.6 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.3 | GO:0051918 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.4 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0030849 | Y chromosome(GO:0000806) autosome(GO:0030849) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0043073 | male germ cell nucleus(GO:0001673) germ cell nucleus(GO:0043073) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |