Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
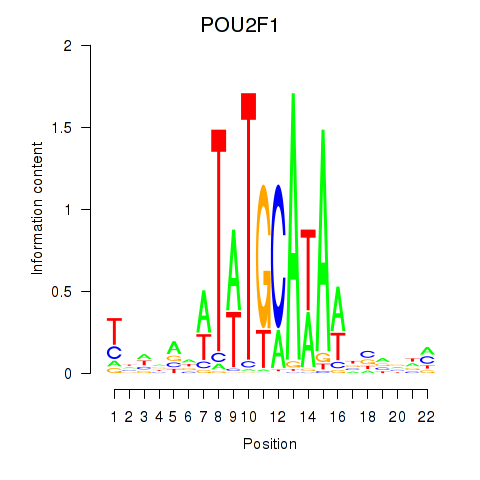
Results for POU2F1
Z-value: 1.09
Transcription factors associated with POU2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F1
|
ENSG00000143190.17 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F1 | hg19_v2_chr1_+_167190066_167190156 | 0.64 | 1.6e-04 | Click! |
Activity profile of POU2F1 motif
Sorted Z-values of POU2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_70861647 | 13.96 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chrX_-_99665262 | 8.18 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr14_-_107078851 | 7.98 |
ENST00000390628.2
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr12_-_58329819 | 5.30 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr1_+_104293028 | 5.00 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr5_+_140227048 | 4.78 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr14_-_21270995 | 4.77 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr14_-_21270561 | 4.71 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_-_148939835 | 4.01 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_104159999 | 3.76 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr14_-_106642049 | 3.71 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr4_-_100356551 | 3.68 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr18_-_24722995 | 3.40 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr6_+_135502408 | 3.33 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_-_100356291 | 3.30 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_+_135502466 | 3.27 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_58329888 | 2.98 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr10_+_91589309 | 2.90 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chrX_+_57313113 | 2.85 |
ENST00000374900.4
|
FAAH2
|
fatty acid amide hydrolase 2 |
| chr10_+_91589261 | 2.85 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr11_-_26588634 | 2.75 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr11_+_6897856 | 2.73 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr15_-_45670924 | 2.63 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr7_+_48211048 | 2.33 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr7_+_90338712 | 2.29 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr10_-_75118471 | 2.12 |
ENST00000340329.3
|
TTC18
|
tetratricopeptide repeat domain 18 |
| chr1_-_223308098 | 2.12 |
ENST00000342210.6
|
TLR5
|
toll-like receptor 5 |
| chr6_-_46922659 | 2.04 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr5_+_125758813 | 2.01 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr4_-_110723194 | 1.94 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr4_-_110723134 | 1.92 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_-_110723335 | 1.91 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr18_-_52989217 | 1.90 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr16_-_18462221 | 1.87 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_26217159 | 1.87 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr10_-_116286563 | 1.84 |
ENST00000369253.2
|
ABLIM1
|
actin binding LIM protein 1 |
| chr5_-_176326333 | 1.83 |
ENST00000292432.5
|
HK3
|
hexokinase 3 (white cell) |
| chr5_+_125758865 | 1.83 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_48866517 | 1.83 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr4_+_74702214 | 1.78 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr3_-_121379739 | 1.65 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr14_-_106453155 | 1.63 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr6_+_135502501 | 1.61 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_7245125 | 1.59 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr5_-_13944652 | 1.56 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr11_-_110561721 | 1.53 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr10_-_116286656 | 1.45 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr8_+_18067602 | 1.44 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr7_-_99149715 | 1.32 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr11_+_120107344 | 1.27 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr3_+_32737027 | 1.26 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr9_+_108424738 | 1.24 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr1_-_213020991 | 1.22 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr16_+_19222479 | 1.19 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr4_+_146539415 | 1.19 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr3_+_185046676 | 1.17 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr19_+_56368803 | 1.15 |
ENST00000587891.1
|
NLRP4
|
NLR family, pyrin domain containing 4 |
| chr8_-_33370607 | 1.14 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr8_+_86121448 | 1.11 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr15_-_89089860 | 1.10 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr5_-_111092873 | 1.09 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr1_-_89736434 | 1.07 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr6_-_152489484 | 0.98 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_111093081 | 0.95 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr19_+_17337007 | 0.94 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr12_-_111358372 | 0.93 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr19_+_17337027 | 0.90 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr2_-_197226875 | 0.88 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_196621002 | 0.88 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr1_-_101360331 | 0.86 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr19_-_44124019 | 0.83 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr6_+_122720681 | 0.82 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr12_+_40618873 | 0.81 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr13_+_108870714 | 0.78 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr9_+_124088860 | 0.78 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr2_-_111334678 | 0.78 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr7_+_55433131 | 0.76 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr3_-_155524049 | 0.74 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr6_+_147527103 | 0.74 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr3_+_149530836 | 0.73 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr19_+_58258164 | 0.73 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr17_-_37844267 | 0.72 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr4_-_144940477 | 0.71 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr2_+_110551851 | 0.69 |
ENST00000272454.6
ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5
|
RANBP2-like and GRIP domain containing 5 |
| chr9_+_120466650 | 0.66 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chrX_-_131262048 | 0.64 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chrX_-_34150428 | 0.63 |
ENST00000346193.3
|
FAM47A
|
family with sequence similarity 47, member A |
| chr17_+_37844331 | 0.62 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr3_-_3221358 | 0.57 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr1_-_157567868 | 0.57 |
ENST00000271532.1
|
FCRL4
|
Fc receptor-like 4 |
| chr6_-_15586238 | 0.57 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr16_-_46782221 | 0.55 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr20_+_5731083 | 0.55 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr2_-_113191096 | 0.55 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr19_-_44123734 | 0.53 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr9_-_140196703 | 0.52 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chrX_+_119384607 | 0.52 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr10_-_52008313 | 0.52 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr1_+_6615241 | 0.49 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr7_+_33765593 | 0.47 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr9_-_123812542 | 0.47 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr9_-_21335356 | 0.46 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr1_+_161736072 | 0.45 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr7_+_64838712 | 0.43 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr9_+_120466610 | 0.42 |
ENST00000394487.4
|
TLR4
|
toll-like receptor 4 |
| chr2_+_109237717 | 0.40 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_43239231 | 0.40 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_+_135388147 | 0.39 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr16_+_16434185 | 0.39 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr14_+_21249200 | 0.36 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr3_+_157154578 | 0.36 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr1_+_100436065 | 0.35 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr7_+_64838786 | 0.35 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr14_-_68283291 | 0.35 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chr11_-_14521379 | 0.33 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr13_+_52436111 | 0.32 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr17_-_7387524 | 0.32 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr13_+_111855414 | 0.31 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_+_28562617 | 0.31 |
ENST00000497986.1
ENST00000335514.5 ENST00000468425.2 ENST00000465645.1 |
ATPIF1
|
ATPase inhibitory factor 1 |
| chr5_+_61874562 | 0.31 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr2_+_219524379 | 0.30 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr5_+_67588391 | 0.30 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_+_129083772 | 0.30 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr4_+_120056939 | 0.30 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr20_+_39657454 | 0.29 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr12_-_72057571 | 0.29 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr3_+_118892362 | 0.29 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chrX_-_6146876 | 0.29 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr12_+_80750134 | 0.27 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chr10_+_13628921 | 0.26 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr6_+_127898312 | 0.26 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_-_241683001 | 0.26 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr1_-_160681593 | 0.25 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr10_+_13628933 | 0.24 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr1_-_169703203 | 0.24 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr16_-_20416053 | 0.23 |
ENST00000302451.4
|
PDILT
|
protein disulfide isomerase-like, testis expressed |
| chr18_+_3252206 | 0.22 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_103035102 | 0.22 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chrX_-_19988382 | 0.21 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chrX_+_154254960 | 0.20 |
ENST00000369498.3
|
FUNDC2
|
FUN14 domain containing 2 |
| chr12_-_127174845 | 0.20 |
ENST00000540814.1
ENST00000541065.1 ENST00000541769.1 |
RP11-407A16.3
RP11-407A16.4
|
RP11-407A16.3 RP11-407A16.4 |
| chr19_-_14682838 | 0.20 |
ENST00000215565.2
|
NDUFB7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa |
| chr3_-_149293990 | 0.18 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_44001172 | 0.17 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr9_-_116065551 | 0.17 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr2_+_169658928 | 0.17 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr12_+_72056773 | 0.16 |
ENST00000308086.2
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr11_+_85359062 | 0.16 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr11_-_5207612 | 0.15 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr5_-_133510456 | 0.15 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr2_+_169659121 | 0.15 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr6_-_132834184 | 0.15 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr12_-_118796910 | 0.15 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_87135076 | 0.15 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr1_-_6614565 | 0.15 |
ENST00000377705.5
|
NOL9
|
nucleolar protein 9 |
| chr12_-_127174806 | 0.14 |
ENST00000545853.1
ENST00000537478.1 |
RP11-407A16.3
|
RP11-407A16.3 |
| chr22_+_23029188 | 0.13 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr17_-_29624343 | 0.13 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr8_-_86290333 | 0.12 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr1_-_226926864 | 0.12 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr5_+_150827143 | 0.11 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr2_+_36923830 | 0.10 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr21_-_33975547 | 0.10 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_26033796 | 0.09 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr5_+_112227311 | 0.09 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr3_-_165555200 | 0.09 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_-_35230649 | 0.09 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chr22_+_41697520 | 0.08 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr14_-_65346555 | 0.08 |
ENST00000542895.1
ENST00000556626.1 |
SPTB
|
spectrin, beta, erythrocytic |
| chr10_+_126630692 | 0.07 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chr11_-_62609281 | 0.06 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr5_+_140739537 | 0.06 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr2_-_89459813 | 0.05 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr17_+_34391625 | 0.05 |
ENST00000004921.3
|
CCL18
|
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| chr2_-_152684977 | 0.05 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chrX_-_130423386 | 0.04 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr17_+_75181292 | 0.04 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr10_-_31288398 | 0.04 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr17_-_55822653 | 0.03 |
ENST00000299415.2
|
AC007431.1
|
coiled-coil domain containing 182 |
| chr6_-_13621126 | 0.03 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chrX_-_130423200 | 0.03 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr9_-_21202204 | 0.03 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr1_-_106161540 | 0.02 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr12_-_91546926 | 0.02 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_-_11779174 | 0.02 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr16_-_90142338 | 0.02 |
ENST00000407825.1
ENST00000449207.2 |
PRDM7
|
PR domain containing 7 |
| chrX_-_130423240 | 0.01 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr4_-_100065440 | 0.01 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr18_+_5748793 | 0.01 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr5_+_126626498 | 0.00 |
ENST00000503335.2
ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10
|
multiple EGF-like-domains 10 |
| chr6_-_11779014 | 0.00 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.3 | 7.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.9 | 14.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.7 | 2.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.5 | 1.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.5 | 3.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 1.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.3 | 1.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 2.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.8 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 0.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.9 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.5 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.2 | 0.8 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 4.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 8.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 9.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 1.0 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 3.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.6 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 12.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 6.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 1.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.3 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.8 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 4.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 2.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.3 | GO:0097105 | brainstem development(GO:0003360) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 1.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.9 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 1.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 0.8 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 8.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.5 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 7.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 5.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.7 | 7.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 1.4 | 9.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.9 | 3.8 | GO:0016160 | amylase activity(GO:0016160) |
| 0.7 | 8.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 3.4 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.4 | 4.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 1.8 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.9 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.2 | 8.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 4.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 13.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 4.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 6.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 6.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |