Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
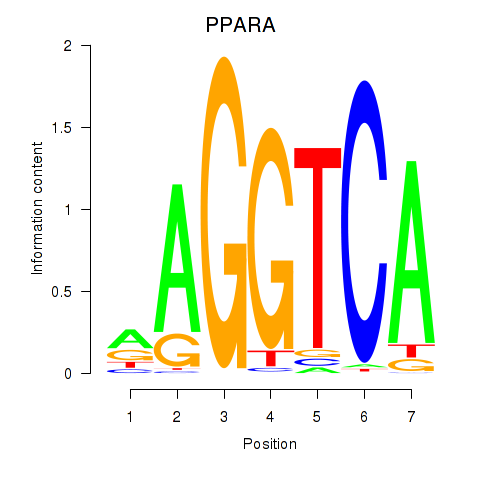
Results for PPARA
Z-value: 1.11
Transcription factors associated with PPARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARA
|
ENSG00000186951.12 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARA | hg19_v2_chr22_+_46546494_46546525 | -0.46 | 1.0e-02 | Click! |
Activity profile of PPARA motif
Sorted Z-values of PPARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63638339 | 10.92 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr3_-_45957088 | 8.54 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_45957534 | 6.65 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr12_-_25348007 | 6.21 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr16_-_67427389 | 6.03 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_-_34381536 | 5.85 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr9_-_34381511 | 5.79 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr15_+_71145578 | 5.77 |
ENST00000544974.2
ENST00000558546.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_28113217 | 4.54 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr9_+_34458771 | 4.52 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr1_-_104238912 | 4.40 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_-_104239076 | 4.36 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr3_-_9994021 | 3.72 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr2_-_27712583 | 3.71 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr4_+_15471489 | 3.65 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr15_+_43809797 | 3.50 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr9_-_117150243 | 3.42 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr14_-_21492251 | 3.41 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492113 | 3.39 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr16_+_777118 | 3.33 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr11_-_111175739 | 3.25 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr15_+_82555125 | 3.24 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr20_+_31755934 | 3.14 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr1_+_104159999 | 3.09 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr17_+_4981535 | 2.97 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr22_+_31518938 | 2.74 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_46640750 | 2.72 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr19_+_35629702 | 2.58 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr7_+_116660246 | 2.45 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr12_+_6930813 | 2.41 |
ENST00000428545.2
|
GPR162
|
G protein-coupled receptor 162 |
| chr12_+_6930703 | 2.38 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr17_+_40440481 | 2.31 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr3_+_184055240 | 2.30 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr5_+_156712372 | 2.26 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_+_68071389 | 2.24 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_68071458 | 2.23 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_-_6433765 | 2.19 |
ENST00000321510.6
|
SLC25A41
|
solute carrier family 25, member 41 |
| chr1_-_160231451 | 2.15 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr17_+_53344945 | 2.04 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr1_+_54359854 | 2.03 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr11_-_108408895 | 2.03 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr7_+_121513143 | 1.99 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr4_+_30721968 | 1.99 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr1_-_226129083 | 1.96 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr8_+_104831554 | 1.96 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_226129189 | 1.92 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr4_+_41614909 | 1.89 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_+_104178946 | 1.89 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr22_+_32149927 | 1.89 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr1_+_212738676 | 1.83 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr12_+_53817633 | 1.79 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr3_-_52486841 | 1.79 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr3_+_160559931 | 1.71 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr2_+_230787213 | 1.69 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr16_+_53133070 | 1.69 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_133772794 | 1.68 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr1_-_217250231 | 1.66 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr2_-_207024233 | 1.66 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_+_230787201 | 1.64 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr6_+_39760129 | 1.60 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr6_+_143999072 | 1.60 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr2_-_25194963 | 1.58 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr15_+_76352178 | 1.58 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr4_+_106067943 | 1.56 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr8_-_108510224 | 1.56 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr3_-_114343768 | 1.54 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_+_14444063 | 1.53 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr19_-_50311896 | 1.51 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr12_+_6930964 | 1.51 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr19_+_13134772 | 1.50 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chrX_-_135849484 | 1.49 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_+_72204755 | 1.49 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr11_-_86383157 | 1.47 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr3_-_114866084 | 1.47 |
ENST00000357258.3
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_-_73687997 | 1.46 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr16_+_28303804 | 1.46 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr11_+_120894781 | 1.46 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr16_+_776936 | 1.44 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_-_84273304 | 1.42 |
ENST00000308251.4
ENST00000568181.1 |
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chrX_-_108868390 | 1.42 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr11_-_8832182 | 1.41 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr5_+_140027355 | 1.41 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr11_-_86383370 | 1.40 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr14_-_31889782 | 1.40 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr10_+_76586348 | 1.40 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr5_+_133451254 | 1.40 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr5_+_139927213 | 1.38 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr22_-_24110063 | 1.37 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr10_+_81107271 | 1.35 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr9_+_115983808 | 1.34 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr6_-_87804815 | 1.34 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr20_+_53092123 | 1.33 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr16_-_67700594 | 1.31 |
ENST00000602644.1
ENST00000243878.4 |
ENKD1
|
enkurin domain containing 1 |
| chr8_+_110346546 | 1.30 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr1_+_9648921 | 1.29 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr15_+_59730348 | 1.29 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr4_+_30723003 | 1.28 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr12_-_48744554 | 1.28 |
ENST00000544117.2
ENST00000548932.1 ENST00000549125.1 ENST00000301042.3 ENST00000547026.1 |
ZNF641
|
zinc finger protein 641 |
| chr16_-_28937027 | 1.24 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr1_-_160232312 | 1.24 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr2_+_217498105 | 1.22 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_+_107349392 | 1.19 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr11_-_66445219 | 1.18 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr8_+_37553261 | 1.17 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr10_-_104178857 | 1.15 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr12_-_9268707 | 1.15 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr2_-_219433014 | 1.14 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr6_+_44310376 | 1.14 |
ENST00000515220.1
ENST00000323108.8 |
SPATS1
|
spermatogenesis associated, serine-rich 1 |
| chr12_+_56477093 | 1.14 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_73109339 | 1.14 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr7_+_94536898 | 1.12 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_+_58755184 | 1.11 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr5_-_169626104 | 1.10 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr16_+_30710462 | 1.10 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr5_-_140027175 | 1.09 |
ENST00000512088.1
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr13_+_103046954 | 1.08 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr19_+_16771936 | 1.06 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr14_-_21491477 | 1.06 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr1_-_217311090 | 1.05 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr18_-_12377283 | 1.05 |
ENST00000269143.3
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr12_+_109577202 | 1.05 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_-_7293942 | 1.05 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr17_-_9940058 | 1.05 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chr2_+_201936458 | 1.04 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr2_+_219135115 | 1.03 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr1_+_222988406 | 1.03 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr17_-_46035187 | 1.03 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr10_-_94050820 | 1.02 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_114843889 | 1.01 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr4_-_103682145 | 1.00 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr1_+_159750776 | 1.00 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr17_+_38975358 | 1.00 |
ENST00000436612.1
ENST00000301665.3 |
TMEM99
|
transmembrane protein 99 |
| chr1_+_159750720 | 1.00 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr14_-_24911448 | 0.99 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr1_+_169075554 | 0.99 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr9_-_33402506 | 0.99 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr5_-_42811986 | 0.98 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_+_222988464 | 0.98 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr14_+_23352374 | 0.97 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr12_-_46121554 | 0.97 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr9_+_95997205 | 0.97 |
ENST00000411624.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr7_-_73153161 | 0.97 |
ENST00000395147.4
|
ABHD11
|
abhydrolase domain containing 11 |
| chr15_-_42783303 | 0.97 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr11_+_3876859 | 0.96 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr19_-_55881741 | 0.96 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr1_+_16062820 | 0.96 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chrX_-_102531717 | 0.95 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr9_+_1051481 | 0.95 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr17_-_38256973 | 0.95 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr16_+_28875126 | 0.94 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_174992544 | 0.93 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr12_-_56727676 | 0.91 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_+_179322481 | 0.91 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr6_+_149068464 | 0.91 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr20_-_44485835 | 0.90 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr11_-_110583912 | 0.89 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr1_+_223101757 | 0.89 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr7_-_73153122 | 0.89 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr16_+_691792 | 0.88 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr1_+_222988363 | 0.88 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr1_-_185286461 | 0.88 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr5_-_140027357 | 0.88 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr22_+_47158518 | 0.87 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr16_+_2588012 | 0.87 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr1_-_157108266 | 0.87 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chrX_-_15619076 | 0.86 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_-_46000064 | 0.86 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr14_-_24911868 | 0.86 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr10_-_104179682 | 0.86 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_+_160097462 | 0.86 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr7_+_150065879 | 0.85 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr22_-_39239987 | 0.85 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr5_-_42812143 | 0.85 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_-_150777874 | 0.84 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr4_+_41614720 | 0.84 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_+_88852059 | 0.84 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr10_-_106240032 | 0.84 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr17_-_7123021 | 0.84 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr2_-_233877912 | 0.84 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chrX_+_48681768 | 0.84 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr14_-_89021077 | 0.83 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr4_+_26322409 | 0.83 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_-_10979750 | 0.83 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr7_-_91875358 | 0.82 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr1_+_19923454 | 0.82 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr16_-_4466565 | 0.82 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr11_+_111169565 | 0.82 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr14_+_73525144 | 0.82 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chrX_+_152990302 | 0.81 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr12_+_56414795 | 0.81 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr16_+_57653989 | 0.81 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_+_7618413 | 0.80 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_+_174769006 | 0.80 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_72936608 | 0.77 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_-_31938700 | 0.77 |
ENST00000495340.1
|
DXO
|
decapping exoribonuclease |
| chr14_+_58765103 | 0.77 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_35756186 | 0.77 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr21_-_43816052 | 0.76 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr16_-_88717423 | 0.76 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr6_+_35227449 | 0.76 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.9 | 4.5 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.7 | 2.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.6 | 1.8 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.6 | 1.7 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.6 | 2.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.5 | 3.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 7.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.5 | 1.5 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 1.8 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.4 | 1.8 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 3.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.4 | 2.6 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 5.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.4 | 3.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.4 | 1.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.4 | 2.6 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 | 1.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 1.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 3.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 0.9 | GO:0060086 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.3 | 0.9 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 1.1 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.2 | 2.0 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.7 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 1.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 2.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.2 | 4.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.8 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 | 1.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 0.6 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.2 | 0.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.8 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.3 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 0.7 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 0.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 0.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 0.5 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 0.5 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.2 | 2.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 1.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 2.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.7 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.9 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 3.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 2.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 1.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.4 | GO:0021966 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance(GO:0021966) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.6 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 8.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:1903947 | regulation of heart looping(GO:1901207) positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.6 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 1.6 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.9 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.5 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 0.8 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.2 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 1.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 1.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.8 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.3 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 2.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 1.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.5 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.6 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.2 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.1 | 4.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 1.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 2.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.3 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.5 | GO:0035900 | isocitrate metabolic process(GO:0006102) response to isolation stress(GO:0035900) |
| 0.1 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.3 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 5.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 1.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 2.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.8 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 2.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) surfactant homeostasis(GO:0043129) |
| 0.0 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 2.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 2.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 1.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.8 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0060999 | regulation of dendritic spine development(GO:0060998) positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 2.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 2.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 2.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 4.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) regulation of alcohol biosynthetic process(GO:1902930) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:0035966 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.5 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.6 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.7 | 2.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 1.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.5 | 5.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 4.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 1.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.2 | 1.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 3.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 4.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.5 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 0.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.6 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 3.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 7.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 4.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 3.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 5.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 4.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.0 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 2.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.7 | 2.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.7 | 2.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.5 | 2.9 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 1.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 2.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 1.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.4 | 1.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 3.4 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.3 | 0.9 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.3 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 0.8 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 4.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 0.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 0.9 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.2 | 0.8 | GO:0004040 | amidase activity(GO:0004040) |
| 0.2 | 0.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 2.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 4.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 1.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 1.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 0.5 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 2.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.7 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 1.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 8.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 4.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.6 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 1.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.5 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 2.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.9 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.8 | GO:0050664 | superoxide-generating NADPH oxidase activity(GO:0016175) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 3.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 5.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.2 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 3.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 5.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 9.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |