Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
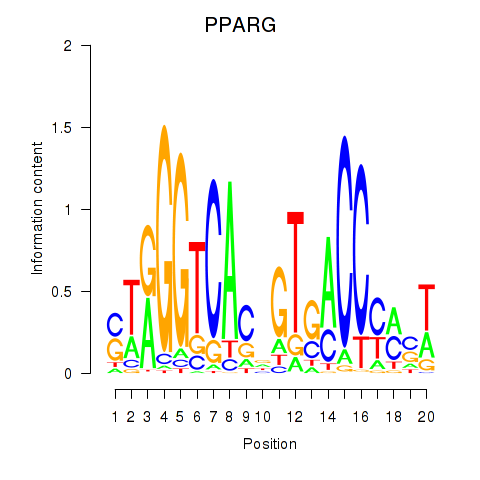
Results for PPARG
Z-value: 0.61
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | peroxisome proliferator activated receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12329358_12329393, hg19_v2_chr3_+_12329397_12329433, hg19_v2_chr3_+_12392971_12392983, hg19_v2_chr3_+_12330560_12330579 | -0.39 | 3.1e-02 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_31870927 | 2.63 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr2_+_120187465 | 1.41 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr20_+_37434329 | 1.15 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr19_+_49661037 | 1.04 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_50016411 | 0.99 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_-_21289627 | 0.92 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr8_-_143857402 | 0.92 |
ENST00000523332.1
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr19_+_49661079 | 0.90 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr7_-_99569468 | 0.89 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_+_67218124 | 0.87 |
ENST00000282670.2
|
TCTEX1D1
|
Tctex1 domain containing 1 |
| chr1_-_223853348 | 0.85 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr18_+_5238055 | 0.83 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr1_+_217804661 | 0.81 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr19_+_56717283 | 0.80 |
ENST00000376267.1
|
ZSCAN5C
|
zinc finger and SCAN domain containing 5C |
| chr19_+_49660997 | 0.78 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr15_+_71145578 | 0.78 |
ENST00000544974.2
ENST00000558546.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_153433120 | 0.73 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr8_-_101348408 | 0.71 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr15_+_65204075 | 0.69 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr14_-_106642049 | 0.68 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr19_+_50016610 | 0.66 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr22_+_39853258 | 0.66 |
ENST00000341184.6
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr10_+_114135952 | 0.65 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr1_-_47407097 | 0.62 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr3_-_122512619 | 0.57 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr9_-_117150243 | 0.57 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr6_-_24911195 | 0.55 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr14_-_106330458 | 0.54 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr17_-_41984835 | 0.54 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chrX_+_9433048 | 0.53 |
ENST00000217964.7
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr17_+_48172639 | 0.53 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr6_-_43478239 | 0.49 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr1_+_20512568 | 0.48 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr11_-_5255696 | 0.47 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr14_+_105953204 | 0.46 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr1_+_153388993 | 0.45 |
ENST00000368729.4
|
S100A7A
|
S100 calcium binding protein A7A |
| chr6_-_4079334 | 0.45 |
ENST00000492651.1
ENST00000498677.1 ENST00000274673.3 |
FAM217A
|
family with sequence similarity 217, member A |
| chr11_-_5255861 | 0.45 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr14_+_105953246 | 0.44 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr19_+_48958766 | 0.44 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr5_+_156696362 | 0.43 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_125687987 | 0.42 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_-_145968923 | 0.42 |
ENST00000493382.1
ENST00000354952.2 ENST00000383083.2 |
PLSCR4
|
phospholipid scramblase 4 |
| chr1_-_46642154 | 0.41 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr10_+_70587279 | 0.39 |
ENST00000298596.6
ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1
|
storkhead box 1 |
| chr22_-_21579843 | 0.38 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr11_+_1942580 | 0.38 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chrX_+_133371077 | 0.37 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr3_+_8775466 | 0.36 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr16_+_67022476 | 0.36 |
ENST00000540947.2
|
CES4A
|
carboxylesterase 4A |
| chr16_+_67022582 | 0.35 |
ENST00000541479.1
ENST00000338718.4 |
CES4A
|
carboxylesterase 4A |
| chr4_-_1670632 | 0.35 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr3_-_145968857 | 0.34 |
ENST00000433593.2
ENST00000476202.1 ENST00000460885.1 |
PLSCR4
|
phospholipid scramblase 4 |
| chr8_-_145115584 | 0.34 |
ENST00000426825.1
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing) |
| chr17_-_40346477 | 0.34 |
ENST00000593209.1
ENST00000587427.1 ENST00000588352.1 ENST00000414034.3 ENST00000590249.1 |
GHDC
|
GH3 domain containing |
| chr16_+_67022633 | 0.34 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr2_-_233792837 | 0.33 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr6_+_133562744 | 0.33 |
ENST00000525849.1
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr12_+_122181529 | 0.31 |
ENST00000541467.1
|
TMEM120B
|
transmembrane protein 120B |
| chr2_+_112895939 | 0.30 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr19_-_55658687 | 0.30 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr18_+_5238549 | 0.29 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr19_+_40093173 | 0.29 |
ENST00000600141.1
|
LGALS13
|
lectin, galactoside-binding, soluble, 13 |
| chr14_-_106453155 | 0.29 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr16_+_476379 | 0.29 |
ENST00000434585.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr7_+_75511362 | 0.28 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr10_-_28623368 | 0.28 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr11_+_75479763 | 0.28 |
ENST00000228027.7
|
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr11_+_61248583 | 0.28 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr11_+_46740730 | 0.28 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr19_-_7698599 | 0.27 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr16_-_67969888 | 0.26 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr2_+_9778872 | 0.26 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr6_+_46097711 | 0.26 |
ENST00000321037.4
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) |
| chr14_+_21152259 | 0.25 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr7_+_150498783 | 0.25 |
ENST00000475536.1
ENST00000468689.1 |
TMEM176A
|
transmembrane protein 176A |
| chr7_+_150498610 | 0.25 |
ENST00000461345.1
|
TMEM176A
|
transmembrane protein 176A |
| chr22_+_23229960 | 0.25 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr22_+_22681656 | 0.25 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr22_-_23484246 | 0.24 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr9_-_130679257 | 0.23 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr7_-_44198874 | 0.23 |
ENST00000395796.3
ENST00000345378.2 |
GCK
|
glucokinase (hexokinase 4) |
| chr19_+_44645700 | 0.23 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr4_-_25865159 | 0.23 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr8_+_37553261 | 0.23 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr19_+_4343691 | 0.23 |
ENST00000597036.1
|
MPND
|
MPN domain containing |
| chr19_+_45349630 | 0.22 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr5_-_180076580 | 0.22 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr17_-_38821373 | 0.22 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_-_20446020 | 0.22 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr18_+_55711575 | 0.21 |
ENST00000356462.6
ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_+_109915422 | 0.21 |
ENST00000280774.5
ENST00000536398.1 ENST00000539599.1 ENST00000342494.3 ENST00000340074.5 ENST00000540230.1 ENST00000537063.1 |
UBE3B
|
ubiquitin protein ligase E3B |
| chr15_+_41056255 | 0.21 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr17_+_77704681 | 0.21 |
ENST00000328313.5
|
ENPP7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr6_+_31783291 | 0.21 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr15_+_41056218 | 0.21 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr22_+_50312316 | 0.21 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr3_+_48507210 | 0.21 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr19_+_1491144 | 0.20 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chr11_+_308143 | 0.20 |
ENST00000399817.4
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr19_+_52800410 | 0.20 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr19_-_7697857 | 0.19 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr1_-_109203997 | 0.19 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr11_-_65686496 | 0.19 |
ENST00000449692.3
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr11_-_6440283 | 0.19 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_+_4343584 | 0.19 |
ENST00000596722.1
|
MPND
|
MPN domain containing |
| chr22_+_50312274 | 0.19 |
ENST00000404488.3
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr14_+_100848311 | 0.19 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr1_+_170501270 | 0.19 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr9_+_95997205 | 0.18 |
ENST00000411624.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr3_-_15643090 | 0.18 |
ENST00000451445.2
ENST00000421993.1 |
HACL1
|
2-hydroxyacyl-CoA lyase 1 |
| chr8_-_134115118 | 0.18 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr11_-_1593150 | 0.18 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr13_+_107029084 | 0.18 |
ENST00000444865.1
|
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr12_+_113796347 | 0.18 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr21_-_33984888 | 0.18 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr3_-_123710893 | 0.18 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr3_-_15643060 | 0.18 |
ENST00000414979.1
ENST00000435217.2 ENST00000456194.2 ENST00000457447.2 |
HACL1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_-_179112189 | 0.18 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr21_-_40817645 | 0.18 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr16_+_75182376 | 0.18 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr21_-_33984865 | 0.18 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr19_-_22018966 | 0.17 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr1_-_109203685 | 0.17 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr8_+_11141925 | 0.17 |
ENST00000221086.3
|
MTMR9
|
myotubularin related protein 9 |
| chr17_-_56032602 | 0.17 |
ENST00000577840.1
|
CUEDC1
|
CUE domain containing 1 |
| chr21_-_33985127 | 0.17 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr11_+_47587129 | 0.17 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr1_-_161059380 | 0.17 |
ENST00000368012.3
|
PVRL4
|
poliovirus receptor-related 4 |
| chr9_+_136243264 | 0.17 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr19_-_55658281 | 0.17 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr12_-_53601000 | 0.16 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr3_+_48507621 | 0.16 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr21_+_47063590 | 0.16 |
ENST00000400314.1
|
PCBP3
|
poly(rC) binding protein 3 |
| chr12_-_53729525 | 0.16 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr2_-_74779744 | 0.16 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr1_-_40137710 | 0.16 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr3_-_108836977 | 0.16 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr11_-_65686586 | 0.16 |
ENST00000438576.2
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr6_+_31795506 | 0.16 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr11_+_59807748 | 0.16 |
ENST00000278855.2
ENST00000532905.1 |
PLAC1L
|
oocyte secreted protein 2 |
| chr9_+_136243117 | 0.16 |
ENST00000426926.2
ENST00000371957.3 |
C9orf96
|
chromosome 9 open reading frame 96 |
| chr1_-_11115877 | 0.16 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr1_+_6484829 | 0.16 |
ENST00000377828.1
|
ESPN
|
espin |
| chr17_+_55173933 | 0.16 |
ENST00000539273.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr6_-_112575838 | 0.15 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr10_+_695888 | 0.15 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chrX_+_152907913 | 0.15 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr15_-_89089860 | 0.15 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr8_+_11351494 | 0.15 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr17_+_55162453 | 0.15 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr12_-_55375622 | 0.15 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr13_+_33160553 | 0.15 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr4_-_143395551 | 0.15 |
ENST00000507861.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr19_-_45826125 | 0.15 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr19_-_51071302 | 0.15 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr14_-_50319482 | 0.15 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr17_-_1733114 | 0.15 |
ENST00000305513.7
|
SMYD4
|
SET and MYND domain containing 4 |
| chr15_-_83316711 | 0.15 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr7_-_10979750 | 0.14 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr21_-_46293644 | 0.14 |
ENST00000330938.3
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr2_-_74780176 | 0.14 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr9_+_86238016 | 0.14 |
ENST00000530832.1
ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr16_+_85646891 | 0.14 |
ENST00000393243.1
|
GSE1
|
Gse1 coiled-coil protein |
| chr12_-_57504069 | 0.14 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr20_+_54933971 | 0.14 |
ENST00000371384.3
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210, member B |
| chr17_-_76975925 | 0.14 |
ENST00000591274.1
ENST00000589906.1 ENST00000591778.1 ENST00000589775.2 ENST00000585407.1 ENST00000262776.3 |
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr1_-_19186176 | 0.14 |
ENST00000375371.3
|
TAS1R2
|
taste receptor, type 1, member 2 |
| chr4_-_156298028 | 0.14 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr21_-_46293586 | 0.14 |
ENST00000445724.2
ENST00000397887.3 |
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr1_+_201798269 | 0.14 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr17_-_7991021 | 0.14 |
ENST00000319144.4
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr19_-_6502304 | 0.14 |
ENST00000540257.1
ENST00000594276.1 ENST00000594075.1 ENST00000600216.1 ENST00000596926.1 |
TUBB4A
|
tubulin, beta 4A class IVa |
| chr19_-_45735138 | 0.14 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr17_+_27573875 | 0.14 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr1_+_203274639 | 0.13 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr3_+_195447738 | 0.13 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr9_+_86237963 | 0.13 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chrX_+_153770421 | 0.13 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_+_207494853 | 0.13 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr17_-_62502639 | 0.13 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr16_+_1203194 | 0.13 |
ENST00000348261.5
ENST00000358590.4 |
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr6_+_97010424 | 0.13 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr12_-_50294033 | 0.12 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr21_-_33984456 | 0.12 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr16_+_57653989 | 0.12 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_770975 | 0.12 |
ENST00000569529.1
ENST00000564000.1 ENST00000219535.3 |
FAM173A
|
family with sequence similarity 173, member A |
| chr12_-_53730147 | 0.12 |
ENST00000536324.2
|
SP7
|
Sp7 transcription factor |
| chr20_-_33999766 | 0.12 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr16_+_85646763 | 0.12 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr18_-_5238525 | 0.12 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr17_+_77681075 | 0.12 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr19_-_51587502 | 0.12 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chr15_-_41166414 | 0.12 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr11_+_65292538 | 0.12 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr1_+_201617264 | 0.12 |
ENST00000367296.4
|
NAV1
|
neuron navigator 1 |
| chr8_+_11351876 | 0.12 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr1_-_1009683 | 0.12 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr19_+_58193388 | 0.12 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr9_+_141107506 | 0.11 |
ENST00000446912.2
|
FAM157B
|
family with sequence similarity 157, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.4 | 1.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 2.6 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 1.7 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.2 | 0.9 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.6 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.4 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.4 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.3 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.3 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.6 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0060702 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0097676 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.3 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.0 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 2.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 1.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.6 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.4 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |