Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
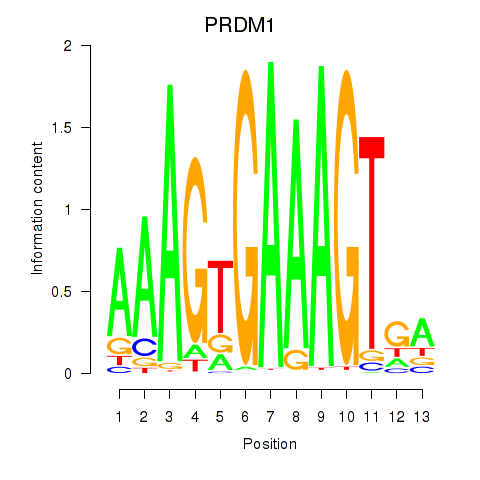
Results for PRDM1
Z-value: 0.58
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106534192_106534224 | -0.45 | 1.2e-02 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29527702 | 3.82 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr6_+_32821924 | 1.50 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_+_39770803 | 1.39 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr17_+_41158742 | 1.21 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr2_+_231191875 | 1.20 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr11_-_47736896 | 1.11 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr8_-_54436491 | 1.09 |
ENST00000426023.1
|
RP11-400K9.4
|
RP11-400K9.4 |
| chr12_+_6561190 | 1.03 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr15_+_71184931 | 1.02 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr22_-_36556821 | 1.00 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr15_+_71185148 | 0.98 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr5_-_96143796 | 0.92 |
ENST00000296754.3
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr1_+_212782012 | 0.89 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr16_-_67970990 | 0.87 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr3_-_49851313 | 0.86 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr4_-_46996424 | 0.84 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr16_+_50775948 | 0.82 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr10_+_91589309 | 0.78 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr3_+_69915385 | 0.77 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_-_122526499 | 0.77 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr2_+_69240302 | 0.69 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr10_+_91589261 | 0.67 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr8_-_23540402 | 0.65 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr15_-_37393406 | 0.65 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr16_+_50775971 | 0.61 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr14_-_21490417 | 0.58 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr11_-_72432950 | 0.57 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_+_32811885 | 0.57 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_-_21490958 | 0.55 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr19_+_13106383 | 0.54 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_+_51318513 | 0.54 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr8_-_29120580 | 0.51 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr14_-_21490653 | 0.51 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr9_-_117568365 | 0.47 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr3_-_114343039 | 0.45 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_-_21490590 | 0.45 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chr17_+_6659153 | 0.45 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr6_+_32811861 | 0.45 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr16_+_50776021 | 0.44 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr17_-_43339474 | 0.44 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr13_+_24144796 | 0.44 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chrX_+_10124977 | 0.43 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_154831779 | 0.43 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr5_+_76506706 | 0.42 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr17_-_43339453 | 0.41 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr13_+_24144509 | 0.41 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr18_-_53070913 | 0.38 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr2_+_234545148 | 0.38 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr4_+_146403912 | 0.37 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr11_+_64323428 | 0.36 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr5_+_92919043 | 0.36 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr1_-_111746966 | 0.36 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr1_-_217250231 | 0.36 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr9_+_84304628 | 0.35 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr7_+_134832808 | 0.35 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr1_+_2985726 | 0.34 |
ENST00000511072.1
ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16
|
PR domain containing 16 |
| chr2_-_231084820 | 0.34 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr5_-_94417339 | 0.34 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr19_+_49977818 | 0.33 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr17_-_56606639 | 0.33 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr17_-_26694979 | 0.33 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr6_-_167275991 | 0.33 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr17_-_56606705 | 0.32 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr6_-_32811771 | 0.32 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_56606664 | 0.32 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr17_-_26695013 | 0.32 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr22_-_31688381 | 0.31 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_+_24605389 | 0.31 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_71710648 | 0.31 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr3_+_185046676 | 0.30 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_231280954 | 0.29 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr11_+_71709938 | 0.29 |
ENST00000393705.4
ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP
|
interleukin 18 binding protein |
| chr8_+_96145974 | 0.28 |
ENST00000315367.3
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr7_-_105332084 | 0.27 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr10_-_112678692 | 0.27 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr1_-_182361327 | 0.27 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr2_-_231084617 | 0.27 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr14_+_24605361 | 0.27 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_-_168864427 | 0.26 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr3_+_158362299 | 0.26 |
ENST00000478576.1
ENST00000264263.5 ENST00000464732.1 |
GFM1
|
G elongation factor, mitochondrial 1 |
| chr7_-_86849025 | 0.26 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr6_-_33282163 | 0.26 |
ENST00000434618.2
ENST00000456592.2 |
TAPBP
|
TAP binding protein (tapasin) |
| chr5_+_112227311 | 0.26 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr5_+_73109339 | 0.25 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr6_-_33281979 | 0.25 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr11_+_71710973 | 0.25 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr5_-_93447333 | 0.25 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr2_-_209010874 | 0.24 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr14_-_24615805 | 0.24 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr10_+_22610124 | 0.24 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr15_-_37392086 | 0.23 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr22_-_31688431 | 0.23 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_-_69229650 | 0.23 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr2_+_69240415 | 0.23 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr1_-_182360918 | 0.23 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr1_+_2985760 | 0.22 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr5_+_17444119 | 0.22 |
ENST00000503267.1
ENST00000504998.1 |
RP11-321E2.4
|
RP11-321E2.4 |
| chr21_+_43933946 | 0.22 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr20_+_30946106 | 0.21 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr2_+_145780725 | 0.21 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_796197 | 0.21 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr2_+_231280908 | 0.20 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr20_+_13976015 | 0.20 |
ENST00000217246.4
|
MACROD2
|
MACRO domain containing 2 |
| chr20_+_388791 | 0.20 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_-_54457783 | 0.20 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr11_-_96076334 | 0.19 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr16_-_3930724 | 0.19 |
ENST00000262367.5
|
CREBBP
|
CREB binding protein |
| chr1_+_110453109 | 0.19 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_46402583 | 0.18 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_-_121655552 | 0.18 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr2_+_179184955 | 0.18 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_-_28220002 | 0.18 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr11_+_46402482 | 0.18 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr16_+_31085714 | 0.17 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr11_+_46402297 | 0.17 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_64008443 | 0.17 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_145780767 | 0.17 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_+_156737292 | 0.17 |
ENST00000271526.4
ENST00000353233.3 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr3_-_46000064 | 0.17 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr6_+_105404899 | 0.17 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr20_-_57089934 | 0.17 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr4_-_74088800 | 0.17 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr2_-_175260368 | 0.17 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr19_-_17186229 | 0.16 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr13_-_95364389 | 0.16 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr12_-_57505121 | 0.16 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_+_64008525 | 0.15 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr14_-_24615523 | 0.15 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr21_+_17909594 | 0.15 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_-_137113604 | 0.14 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr16_-_31085514 | 0.14 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chrX_-_118827113 | 0.14 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr9_-_34710066 | 0.14 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chrX_-_154563889 | 0.14 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr13_+_108921977 | 0.13 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr6_-_167276033 | 0.13 |
ENST00000503859.1
ENST00000506565.1 |
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_-_33282024 | 0.13 |
ENST00000475304.1
ENST00000489157.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr7_-_3083573 | 0.12 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr17_+_7465216 | 0.12 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr17_+_57232690 | 0.12 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr3_+_107241783 | 0.11 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chrX_-_118826784 | 0.10 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr22_+_41697520 | 0.10 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr6_+_146348810 | 0.09 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr2_+_234545092 | 0.09 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_-_153919128 | 0.09 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr3_+_107241882 | 0.08 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr16_+_30675654 | 0.08 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr4_-_111558135 | 0.08 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr1_-_205326022 | 0.07 |
ENST00000367155.3
|
KLHDC8A
|
kelch domain containing 8A |
| chr20_-_18774614 | 0.07 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr1_+_215747118 | 0.07 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr1_-_205326161 | 0.07 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr11_+_46402744 | 0.06 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_146348782 | 0.06 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr2_+_145780739 | 0.06 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr10_-_82049424 | 0.06 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr4_-_146859787 | 0.06 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr1_+_28099683 | 0.06 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chrX_-_118827333 | 0.06 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr1_-_221915418 | 0.05 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr12_+_25205446 | 0.05 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr10_-_21661870 | 0.05 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr2_-_231084659 | 0.04 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr7_-_31380502 | 0.04 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr2_+_234526272 | 0.04 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_9009079 | 0.04 |
ENST00000519699.1
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr10_-_35104185 | 0.04 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr12_-_50561075 | 0.04 |
ENST00000422340.2
ENST00000317551.6 |
CERS5
|
ceramide synthase 5 |
| chr2_+_234590556 | 0.04 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr15_+_85144217 | 0.04 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr12_-_120763739 | 0.04 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr18_-_47017956 | 0.04 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr15_+_75491213 | 0.03 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr21_-_35899113 | 0.03 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr6_-_132910877 | 0.03 |
ENST00000258034.2
|
TAAR5
|
trace amine associated receptor 5 |
| chr13_+_108922228 | 0.03 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_56986768 | 0.03 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr12_+_94656297 | 0.02 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr4_+_186990298 | 0.02 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr12_+_94542459 | 0.02 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chrX_-_139015153 | 0.02 |
ENST00000370557.1
|
ATP11C
|
ATPase, class VI, type 11C |
| chr2_+_175260451 | 0.02 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr1_-_205325850 | 0.02 |
ENST00000537168.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr3_-_167452298 | 0.02 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr6_-_41909191 | 0.02 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr1_-_44497024 | 0.01 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr13_+_102104980 | 0.01 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_+_152020736 | 0.01 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr19_+_24009879 | 0.01 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr2_+_135676381 | 0.01 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr1_+_151372010 | 0.00 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr11_-_40315640 | 0.00 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr20_+_57226841 | 0.00 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr3_+_47422485 | 0.00 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 1.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.3 | 1.0 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.3 | 0.9 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 0.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 1.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 0.6 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 1.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 2.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.7 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.1 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.6 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.8 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 4.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.8 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.7 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 0.9 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 1.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.3 | 0.8 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.2 | 0.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.2 | 3.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 1.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.7 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 3.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |