Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
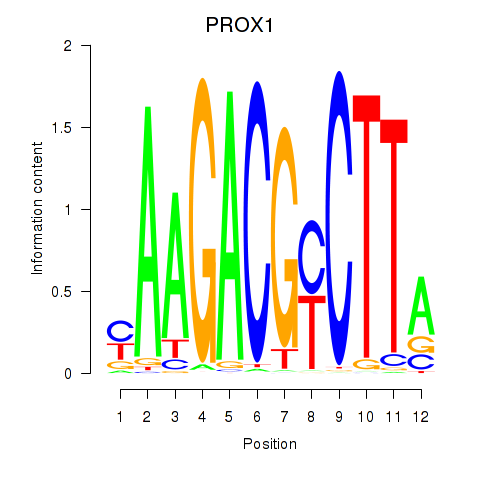
Results for PROX1
Z-value: 0.71
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322 | 0.26 | 1.6e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_22918021 | 5.23 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr20_-_7921090 | 2.50 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr6_-_35992270 | 1.32 |
ENST00000394602.2
ENST00000355574.2 |
SLC26A8
|
solute carrier family 26 (anion exchanger), member 8 |
| chr5_-_156569850 | 1.03 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr17_+_42081914 | 0.88 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr10_-_121296045 | 0.81 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr5_-_157002775 | 0.79 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr15_-_91537723 | 0.78 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr20_+_30028322 | 0.74 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr1_-_202311088 | 0.73 |
ENST00000367274.4
|
UBE2T
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr18_+_2571510 | 0.73 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr7_+_48128316 | 0.72 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr7_+_48128194 | 0.71 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr9_+_132096166 | 0.67 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr2_+_128403439 | 0.61 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr2_+_128403720 | 0.60 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr10_+_14920843 | 0.57 |
ENST00000433779.1
ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr15_-_101835110 | 0.53 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr16_+_50300427 | 0.53 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr9_+_34990219 | 0.52 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_+_16478942 | 0.52 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr1_+_82266053 | 0.51 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr18_-_2571437 | 0.47 |
ENST00000574676.1
ENST00000574538.1 ENST00000319888.6 |
METTL4
|
methyltransferase like 4 |
| chr17_+_26646121 | 0.46 |
ENST00000226230.6
|
TMEM97
|
transmembrane protein 97 |
| chr2_+_97001491 | 0.46 |
ENST00000240423.4
ENST00000427946.1 ENST00000435975.1 ENST00000456906.1 ENST00000455200.1 |
NCAPH
|
non-SMC condensin I complex, subunit H |
| chr1_-_23342340 | 0.44 |
ENST00000566855.1
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr3_-_11888246 | 0.44 |
ENST00000455809.1
ENST00000444133.2 ENST00000273037.5 |
TAMM41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chr7_-_143105941 | 0.42 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr6_-_4079334 | 0.42 |
ENST00000492651.1
ENST00000498677.1 ENST00000274673.3 |
FAM217A
|
family with sequence similarity 217, member A |
| chr15_-_31453157 | 0.41 |
ENST00000559177.1
ENST00000558445.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr6_-_8102279 | 0.40 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_+_89999259 | 0.39 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr14_-_35183755 | 0.39 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr9_+_93564191 | 0.39 |
ENST00000375747.1
|
SYK
|
spleen tyrosine kinase |
| chr1_+_70876891 | 0.38 |
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr18_-_21166841 | 0.38 |
ENST00000269228.5
|
NPC1
|
Niemann-Pick disease, type C1 |
| chr4_-_140477353 | 0.36 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr9_+_75136717 | 0.35 |
ENST00000297784.5
|
TMC1
|
transmembrane channel-like 1 |
| chr12_-_122240792 | 0.35 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr19_-_45909585 | 0.34 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_-_173176452 | 0.34 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr16_+_28722684 | 0.34 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr1_-_111061797 | 0.34 |
ENST00000369771.2
|
KCNA10
|
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr2_-_89247338 | 0.34 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr11_-_61062762 | 0.33 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr1_+_70876926 | 0.33 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr5_-_157002749 | 0.32 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr9_+_125133315 | 0.31 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_+_128886532 | 0.31 |
ENST00000444616.1
ENST00000388795.5 |
C4orf29
|
chromosome 4 open reading frame 29 |
| chr15_-_31453359 | 0.31 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr18_+_33767473 | 0.31 |
ENST00000261326.5
|
MOCOS
|
molybdenum cofactor sulfurase |
| chr3_+_186915274 | 0.30 |
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1 |
| chr19_+_35773242 | 0.30 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr16_+_28722809 | 0.30 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr10_-_92681033 | 0.30 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr16_-_22012419 | 0.29 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr1_-_94050668 | 0.29 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr19_-_46234119 | 0.28 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr19_-_40732594 | 0.28 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr11_-_46408107 | 0.28 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr15_-_76304731 | 0.28 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr1_-_225616515 | 0.28 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr11_+_60609537 | 0.28 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr6_-_32634425 | 0.27 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr21_-_26979786 | 0.27 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr1_+_35544968 | 0.27 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr15_-_74726283 | 0.27 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr14_+_24563262 | 0.26 |
ENST00000559250.1
ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr1_+_165864800 | 0.25 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr10_-_96122682 | 0.25 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr5_-_127418573 | 0.25 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr11_+_28131821 | 0.25 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr1_+_165864821 | 0.25 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr17_+_41994576 | 0.24 |
ENST00000588043.2
|
FAM215A
|
family with sequence similarity 215, member A (non-protein coding) |
| chr17_+_73629500 | 0.23 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr14_+_22458631 | 0.23 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr3_-_122283424 | 0.23 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_-_64739358 | 0.23 |
ENST00000301896.5
ENST00000530444.1 |
C11orf85
|
chromosome 11 open reading frame 85 |
| chr2_-_54087066 | 0.23 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr5_+_131892603 | 0.22 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr12_-_11091862 | 0.22 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr10_+_54074033 | 0.22 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr11_-_57298187 | 0.22 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr10_+_30722866 | 0.22 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_-_47950745 | 0.21 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_-_107235287 | 0.21 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr16_+_57279248 | 0.21 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr3_+_12525931 | 0.20 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr19_+_15752088 | 0.19 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr1_+_241695670 | 0.19 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_-_7307358 | 0.19 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr4_-_159592996 | 0.18 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr9_-_130890662 | 0.18 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chr16_+_4845379 | 0.18 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr16_+_23194033 | 0.18 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr9_+_125132803 | 0.18 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_145826867 | 0.18 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr19_+_45754505 | 0.18 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr17_-_48546324 | 0.17 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr13_-_36788718 | 0.17 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr22_+_24990746 | 0.17 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr12_+_20522179 | 0.17 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr2_+_232826211 | 0.17 |
ENST00000325385.7
ENST00000360410.4 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr11_-_64739542 | 0.17 |
ENST00000536065.1
|
C11orf85
|
chromosome 11 open reading frame 85 |
| chr8_-_41166953 | 0.16 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr3_+_138067314 | 0.16 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr20_+_17949724 | 0.16 |
ENST00000377704.4
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr18_+_60190226 | 0.15 |
ENST00000269499.5
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr11_+_1860200 | 0.15 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr3_-_196014520 | 0.15 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr19_-_16582815 | 0.15 |
ENST00000455140.2
ENST00000248070.6 ENST00000594975.1 |
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr3_+_137906353 | 0.15 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr6_-_42690312 | 0.15 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr5_+_154320623 | 0.15 |
ENST00000523037.1
ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr13_+_49822041 | 0.15 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr19_+_6361754 | 0.15 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr6_+_117198400 | 0.14 |
ENST00000332958.2
|
RFX6
|
regulatory factor X, 6 |
| chr16_+_57279004 | 0.14 |
ENST00000219204.3
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr22_-_39640756 | 0.14 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr1_+_241695424 | 0.14 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_-_109399682 | 0.14 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr14_+_45605157 | 0.13 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr16_+_12070567 | 0.13 |
ENST00000566228.1
|
SNX29
|
sorting nexin 29 |
| chr2_+_232826394 | 0.13 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr11_+_46740730 | 0.13 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr12_-_52946923 | 0.13 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr16_-_10674528 | 0.13 |
ENST00000359543.3
|
EMP2
|
epithelial membrane protein 2 |
| chr1_+_10459111 | 0.13 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr12_-_43945724 | 0.12 |
ENST00000389420.3
ENST00000553158.1 |
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_-_76398077 | 0.12 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr2_+_28718921 | 0.12 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr1_-_204380919 | 0.12 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr1_+_207669573 | 0.12 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr9_-_86571628 | 0.12 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr6_+_29141311 | 0.12 |
ENST00000377167.2
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2 |
| chr5_-_132113083 | 0.11 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr11_+_73882144 | 0.11 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr9_+_131266963 | 0.11 |
ENST00000309971.4
ENST00000372770.4 |
GLE1
|
GLE1 RNA export mediator |
| chr8_+_145064215 | 0.11 |
ENST00000313269.5
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr7_-_12443501 | 0.11 |
ENST00000275358.3
|
VWDE
|
von Willebrand factor D and EGF domains |
| chr17_-_36904437 | 0.11 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr5_-_132113036 | 0.11 |
ENST00000378706.1
|
SEPT8
|
septin 8 |
| chr8_-_87755878 | 0.10 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr18_+_29769978 | 0.10 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr11_-_4719072 | 0.10 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr10_-_69455873 | 0.10 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr22_-_45559540 | 0.10 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr2_+_163175394 | 0.09 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr12_+_54402790 | 0.09 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr12_+_10331605 | 0.09 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr6_-_107235331 | 0.09 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr11_+_73882311 | 0.09 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr22_-_50050986 | 0.09 |
ENST00000405854.1
|
C22orf34
|
chromosome 22 open reading frame 34 |
| chr19_+_58281014 | 0.09 |
ENST00000391702.3
ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586
|
zinc finger protein 586 |
| chr22_+_26138108 | 0.09 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr21_-_40033618 | 0.09 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chrX_+_16668278 | 0.09 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr12_-_66524482 | 0.08 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr19_+_917287 | 0.08 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr8_-_143696833 | 0.08 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr4_+_128886424 | 0.08 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr6_+_28227063 | 0.08 |
ENST00000343684.3
|
NKAPL
|
NFKB activating protein-like |
| chr3_-_33138624 | 0.08 |
ENST00000445488.2
ENST00000307377.8 ENST00000440656.1 ENST00000436768.1 ENST00000307363.5 |
GLB1
|
galactosidase, beta 1 |
| chr5_+_64859066 | 0.08 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr14_-_50559361 | 0.08 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr22_-_45559642 | 0.08 |
ENST00000426282.2
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr2_+_101869262 | 0.08 |
ENST00000289382.3
|
CNOT11
|
CCR4-NOT transcription complex, subunit 11 |
| chr15_+_38746307 | 0.08 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr14_+_45605127 | 0.08 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr17_+_16593539 | 0.08 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr4_-_170947485 | 0.07 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr2_-_89310012 | 0.07 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr4_-_128887069 | 0.07 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr6_+_34725263 | 0.07 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr22_-_32058166 | 0.07 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr4_+_4861385 | 0.07 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr1_-_207226313 | 0.07 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr1_-_15850676 | 0.07 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr11_+_92085262 | 0.07 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr15_-_41047421 | 0.07 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chr1_+_161719552 | 0.07 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr19_-_15529790 | 0.07 |
ENST00000596195.1
ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L
|
A kinase (PRKA) anchor protein 8-like |
| chr3_-_112218205 | 0.07 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr19_-_19754404 | 0.06 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr1_+_150980989 | 0.06 |
ENST00000368935.1
|
PRUNE
|
prune exopolyphosphatase |
| chr6_+_168196210 | 0.06 |
ENST00000597278.1
|
AL009178.1
|
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
| chrX_-_53461305 | 0.06 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr12_+_113229452 | 0.06 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_+_113229543 | 0.06 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr3_-_197025447 | 0.06 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_6361440 | 0.06 |
ENST00000245816.4
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr15_+_81489213 | 0.06 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr3_+_126113734 | 0.06 |
ENST00000352312.1
ENST00000393425.1 |
CCDC37
|
coiled-coil domain containing 37 |
| chr1_+_203830703 | 0.06 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chrY_-_20935572 | 0.06 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr18_+_32073253 | 0.06 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr7_+_128864848 | 0.06 |
ENST00000325006.3
ENST00000446544.2 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_+_6640108 | 0.05 |
ENST00000377674.4
ENST00000488936.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr5_-_132113063 | 0.05 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr12_+_98987369 | 0.05 |
ENST00000401722.3
ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.3 | 1.0 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.3 | 2.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 0.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.4 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.5 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.1 | 0.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.7 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 1.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.6 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0019520 | pentose-phosphate shunt, oxidative branch(GO:0009051) aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.8 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1905176 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.7 | GO:0031262 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 0.1 | 0.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 5.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 1.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 0.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 5.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.5 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 1.4 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |