Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
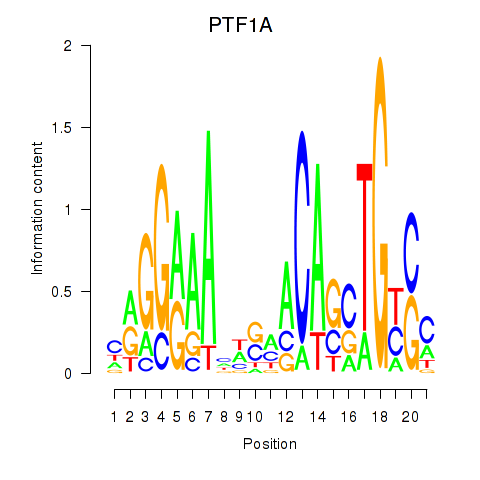
Results for PTF1A
Z-value: 0.67
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_9129598 | 1.22 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr15_-_89764929 | 1.03 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr11_-_111794446 | 0.84 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr8_-_22089845 | 0.79 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_-_22089533 | 0.74 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr1_+_150480576 | 0.72 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr12_+_109826524 | 0.72 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr1_+_150480551 | 0.70 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr20_+_44637526 | 0.66 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr21_+_44073916 | 0.64 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr22_-_50050986 | 0.61 |
ENST00000405854.1
|
C22orf34
|
chromosome 22 open reading frame 34 |
| chr21_+_44073860 | 0.59 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr22_-_50051151 | 0.58 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr11_-_1643368 | 0.58 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr4_+_40198527 | 0.55 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr6_-_46620522 | 0.52 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr2_+_120187465 | 0.51 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr1_-_204654826 | 0.50 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr19_-_46526304 | 0.49 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr8_+_11351494 | 0.48 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr3_-_193272874 | 0.46 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr13_+_113812956 | 0.43 |
ENST00000375547.2
ENST00000342783.4 |
PROZ
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr11_-_1629693 | 0.42 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr19_+_11670245 | 0.41 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr1_-_45956800 | 0.40 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr8_+_11351876 | 0.38 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr5_+_140557371 | 0.37 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr17_+_53342311 | 0.34 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr17_-_43487741 | 0.34 |
ENST00000455881.1
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr7_+_150688166 | 0.34 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr12_-_75603482 | 0.32 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr12_+_1929644 | 0.32 |
ENST00000299194.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr4_+_156587979 | 0.31 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr12_-_75603643 | 0.30 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr22_-_50523760 | 0.30 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr8_-_99306564 | 0.29 |
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr2_-_89278535 | 0.28 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr3_-_52868931 | 0.28 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr8_-_99306611 | 0.28 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chrX_-_21676442 | 0.27 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr1_-_9129735 | 0.26 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr5_+_78532003 | 0.26 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr12_+_1929421 | 0.26 |
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr1_-_9129895 | 0.26 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr10_-_28571015 | 0.26 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_+_173600514 | 0.26 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr10_-_1779663 | 0.26 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr14_-_77737543 | 0.25 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr8_-_125577940 | 0.25 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr3_-_52869205 | 0.25 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr2_+_173600565 | 0.24 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_45687128 | 0.24 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr5_+_173315283 | 0.24 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_-_9129631 | 0.24 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr10_+_99332198 | 0.24 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr21_-_46012386 | 0.23 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr11_-_63933504 | 0.22 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr2_-_228497888 | 0.22 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr19_-_17014408 | 0.22 |
ENST00000594249.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr3_+_40518599 | 0.22 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr1_-_45956822 | 0.22 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chrX_-_107019181 | 0.21 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr6_+_27107053 | 0.21 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr22_+_24999114 | 0.21 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_239140011 | 0.20 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr17_+_40996590 | 0.20 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr9_-_19033237 | 0.20 |
ENST00000380530.1
ENST00000542071.1 |
FAM154A
|
family with sequence similarity 154, member A |
| chr20_-_44259893 | 0.20 |
ENST00000326000.1
|
WFDC9
|
WAP four-disulfide core domain 9 |
| chr9_+_4985228 | 0.20 |
ENST00000381652.3
|
JAK2
|
Janus kinase 2 |
| chr9_-_19033185 | 0.20 |
ENST00000380534.4
|
FAM154A
|
family with sequence similarity 154, member A |
| chr10_-_101841588 | 0.20 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr14_+_77292715 | 0.20 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr20_+_30697298 | 0.19 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr12_+_50451331 | 0.19 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr18_-_5238525 | 0.19 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr7_+_30323923 | 0.19 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr7_-_99097863 | 0.18 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr15_+_41099788 | 0.18 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr16_-_2004683 | 0.17 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr11_+_64052454 | 0.17 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr2_-_160761179 | 0.17 |
ENST00000554112.1
ENST00000553424.1 ENST00000263636.4 ENST00000504764.1 ENST00000505052.1 |
LY75
LY75-CD302
|
lymphocyte antigen 75 LY75-CD302 readthrough |
| chrX_-_48216101 | 0.17 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr2_-_89513402 | 0.17 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_-_71454185 | 0.17 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr12_+_113344755 | 0.17 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_-_152373216 | 0.16 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr11_+_64052692 | 0.16 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chrX_-_134305322 | 0.16 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr1_-_158301312 | 0.15 |
ENST00000368168.3
|
CD1B
|
CD1b molecule |
| chr2_+_85922491 | 0.15 |
ENST00000526018.1
|
GNLY
|
granulysin |
| chr13_-_79979919 | 0.15 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr11_+_64107663 | 0.15 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr3_-_195938256 | 0.15 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr11_+_64052294 | 0.14 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr20_+_42544782 | 0.14 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr1_+_178694300 | 0.14 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_+_183370146 | 0.14 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr4_-_14889791 | 0.14 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr3_-_172241250 | 0.13 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr12_+_31079652 | 0.13 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr6_-_41715128 | 0.13 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr2_+_97426631 | 0.13 |
ENST00000377075.2
|
CNNM4
|
cyclin M4 |
| chr1_+_32084794 | 0.13 |
ENST00000373705.1
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr8_-_135725205 | 0.13 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr14_-_35099315 | 0.13 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr4_-_123843597 | 0.13 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr13_-_79979952 | 0.13 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr6_-_62996066 | 0.13 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr11_+_64052266 | 0.13 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_-_114447683 | 0.12 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_-_114447620 | 0.12 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr2_-_237416181 | 0.12 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr4_-_89744457 | 0.12 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_+_84841242 | 0.12 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr17_+_79859985 | 0.12 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr2_-_237416071 | 0.12 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr19_-_10491130 | 0.12 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr12_-_57644952 | 0.12 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr9_-_104145795 | 0.12 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr5_+_132149017 | 0.11 |
ENST00000378693.2
|
SOWAHA
|
sosondowah ankyrin repeat domain family member A |
| chr6_-_159466042 | 0.11 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr16_+_2802623 | 0.11 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr7_-_142207004 | 0.11 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr2_-_196933536 | 0.11 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr4_-_89744314 | 0.11 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr2_+_44066101 | 0.11 |
ENST00000272286.2
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr5_-_162887071 | 0.11 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr10_+_76871454 | 0.11 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr12_+_113344811 | 0.11 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr4_-_89744365 | 0.11 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr10_-_96829246 | 0.10 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr19_+_17326521 | 0.10 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr11_+_71238313 | 0.10 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr11_-_102576537 | 0.10 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr17_-_3375033 | 0.10 |
ENST00000268981.5
ENST00000397168.3 ENST00000572969.1 ENST00000355380.4 ENST00000571553.1 ENST00000574797.1 ENST00000575375.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr12_-_50677255 | 0.10 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr11_-_61735103 | 0.10 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr3_+_101546827 | 0.10 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr11_+_96123158 | 0.10 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr11_+_7597639 | 0.09 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr18_+_5238055 | 0.09 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr11_+_64058758 | 0.09 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr12_+_112451120 | 0.09 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr3_+_35721182 | 0.09 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr9_-_116061145 | 0.09 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chr17_-_34313685 | 0.09 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr1_-_76076759 | 0.09 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr7_+_7606497 | 0.09 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr10_+_49892904 | 0.08 |
ENST00000360890.2
|
WDFY4
|
WDFY family member 4 |
| chr19_+_54495542 | 0.08 |
ENST00000252729.2
ENST00000352529.1 |
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr4_+_156588115 | 0.08 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr10_-_5855350 | 0.08 |
ENST00000456041.1
ENST00000380181.3 ENST00000418688.1 ENST00000380132.4 ENST00000609712.1 ENST00000380191.4 |
GDI2
|
GDP dissociation inhibitor 2 |
| chr2_-_86422523 | 0.08 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr12_+_53689309 | 0.08 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr6_-_24489842 | 0.08 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_231114795 | 0.08 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr4_+_156587853 | 0.08 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr5_-_162887054 | 0.07 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr9_+_33795533 | 0.07 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr12_+_52281744 | 0.07 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chr11_+_71249071 | 0.07 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr3_-_12883026 | 0.07 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr5_+_56205878 | 0.07 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr4_+_184826418 | 0.07 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr5_-_16916624 | 0.07 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr1_-_114447412 | 0.07 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr6_-_159466136 | 0.06 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr11_+_8704748 | 0.06 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr14_-_35099377 | 0.06 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr9_-_138853156 | 0.06 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr19_-_10491234 | 0.06 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_+_171454659 | 0.05 |
ENST00000367742.3
ENST00000338920.4 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr19_+_42300548 | 0.05 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr16_-_75569068 | 0.05 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr1_+_228337553 | 0.05 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr2_-_237412331 | 0.05 |
ENST00000418802.1
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chrX_-_44402231 | 0.05 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr19_-_11039261 | 0.04 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr6_-_2962331 | 0.04 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr17_-_73892992 | 0.04 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr16_+_2802316 | 0.04 |
ENST00000301740.8
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr17_+_6918064 | 0.04 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr12_+_113344582 | 0.04 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr20_+_48892848 | 0.04 |
ENST00000422459.1
|
RP11-290F20.3
|
RP11-290F20.3 |
| chr11_-_207221 | 0.04 |
ENST00000486280.1
ENST00000332865.6 ENST00000529614.2 ENST00000325147.9 ENST00000410108.1 ENST00000382762.3 |
BET1L
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr11_-_89609185 | 0.04 |
ENST00000329862.6
|
TRIM64B
|
tripartite motif containing 64B |
| chrX_+_15808569 | 0.04 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr19_+_54496132 | 0.04 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr1_+_100503643 | 0.04 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr4_-_175750364 | 0.04 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr1_+_171454639 | 0.04 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr16_+_89696692 | 0.04 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr1_+_32084641 | 0.03 |
ENST00000373706.5
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr16_-_15950868 | 0.03 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr14_+_70346125 | 0.03 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr19_+_19779686 | 0.03 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr2_+_27435179 | 0.03 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr1_+_12185949 | 0.03 |
ENST00000413146.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr3_+_167453493 | 0.02 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_-_89417335 | 0.02 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PTF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 0.5 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.3 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 0.5 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.7 | GO:2001268 | positive regulation of keratinocyte migration(GO:0051549) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 1.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0060752 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.4 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.9 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 1.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 2.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.2 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 1.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 2.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |