Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
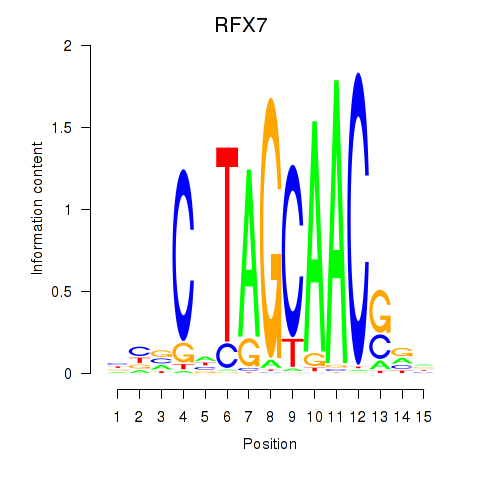
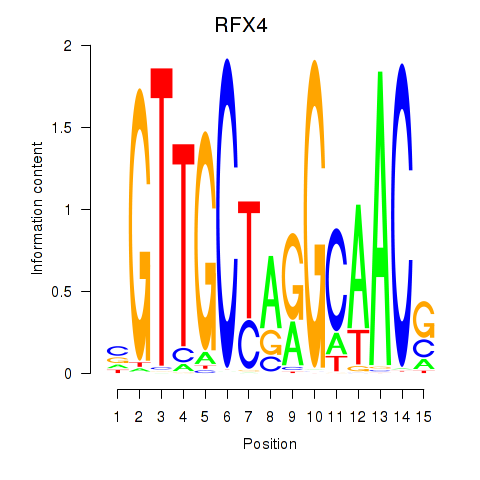
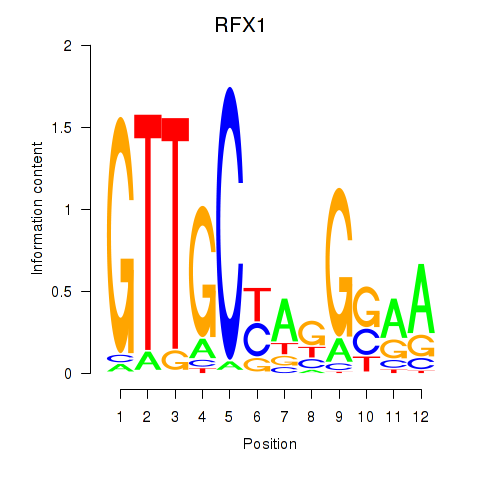
Results for RFX7_RFX4_RFX1
Z-value: 11.26
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.49 | 5.6e-03 | Click! |
| RFX4 | hg19_v2_chr12_+_106976678_106976708, hg19_v2_chr12_+_107078474_107078533 | -0.30 | 1.1e-01 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | -0.25 | 1.8e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_55888186 | 256.66 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr1_-_109655355 | 139.37 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_-_109655377 | 138.59 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_-_161337662 | 134.54 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr9_-_138391692 | 108.68 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr10_+_23216944 | 107.90 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chr10_+_23217006 | 105.22 |
ENST00000376528.4
ENST00000447081.1 |
ARMC3
|
armadillo repeat containing 3 |
| chr16_+_80574854 | 98.84 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr10_-_25305011 | 96.31 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr6_+_163148973 | 94.58 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr20_+_56725952 | 92.44 |
ENST00000371168.3
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr21_-_43916433 | 89.50 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr22_+_23487513 | 85.36 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr22_-_23484246 | 84.24 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr21_-_43916296 | 84.21 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr1_+_85527987 | 82.30 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr1_+_36549676 | 82.23 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr9_-_34381536 | 80.16 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr9_-_34381511 | 79.15 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr6_+_32407619 | 78.41 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr22_-_50970919 | 75.69 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr15_-_55790515 | 74.92 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr17_+_72270429 | 70.55 |
ENST00000311014.6
|
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr17_+_45908974 | 69.99 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr7_-_123174610 | 68.55 |
ENST00000324698.6
ENST00000434450.1 |
IQUB
|
IQ motif and ubiquitin domain containing |
| chr8_+_94767072 | 63.33 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr19_-_50316517 | 61.20 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr6_+_116937636 | 60.81 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr19_-_11545920 | 60.11 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr17_+_72270380 | 59.08 |
ENST00000582036.1
ENST00000307504.5 |
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr1_-_36916066 | 58.95 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr5_-_137475071 | 57.82 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr12_+_111051832 | 55.75 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr12_+_111051902 | 55.05 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr17_+_11501748 | 54.84 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr19_-_50316423 | 54.33 |
ENST00000528094.1
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr19_-_50316489 | 54.00 |
ENST00000533418.1
|
FUZ
|
fuzzy planar cell polarity protein |
| chr2_+_54558004 | 52.91 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr17_+_17876127 | 51.32 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr8_+_94767109 | 50.48 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr11_+_71791693 | 49.53 |
ENST00000289488.2
ENST00000447974.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr2_+_26624775 | 49.46 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr2_-_170550877 | 48.83 |
ENST00000447353.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr10_-_82116505 | 48.77 |
ENST00000372202.1
ENST00000421924.2 ENST00000453477.1 |
DYDC1
|
DPY30 domain containing 1 |
| chrX_+_133371077 | 48.10 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr14_+_96858433 | 47.95 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr10_-_82116497 | 47.88 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr1_-_169396666 | 46.47 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr12_+_119772502 | 45.48 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chrX_-_44202857 | 45.04 |
ENST00000420999.1
|
EFHC2
|
EF-hand domain (C-terminal) containing 2 |
| chr1_-_169396646 | 44.64 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr11_+_71791803 | 43.90 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr1_+_111888890 | 43.76 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr1_+_111889212 | 43.51 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr17_+_42977122 | 43.25 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr1_+_161068179 | 42.99 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr1_-_36915880 | 42.77 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr22_-_50970506 | 42.74 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr11_+_61248583 | 41.99 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr16_-_67700594 | 41.57 |
ENST00000602644.1
ENST00000243878.4 |
ENKD1
|
enkurin domain containing 1 |
| chr22_+_45809560 | 41.13 |
ENST00000342894.3
ENST00000538017.1 |
RIBC2
|
RIB43A domain with coiled-coils 2 |
| chr2_+_29204161 | 40.22 |
ENST00000379558.4
ENST00000403861.2 |
FAM179A
|
family with sequence similarity 179, member A |
| chr9_+_72435709 | 39.92 |
ENST00000377197.3
ENST00000527647.1 |
C9orf135
|
chromosome 9 open reading frame 135 |
| chr3_+_63638339 | 39.77 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr3_-_19975665 | 39.12 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr3_+_97483572 | 38.73 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr11_+_71791359 | 38.71 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr16_+_57406368 | 38.58 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr1_-_36916011 | 38.14 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr2_-_196933536 | 37.42 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr3_+_97483366 | 37.16 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr4_-_177116772 | 37.00 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr2_-_220110187 | 36.30 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_-_220110111 | 36.29 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr10_+_15001430 | 36.05 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr19_+_5914213 | 35.88 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr9_+_34458771 | 35.74 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr12_+_49297899 | 35.31 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr1_+_217804661 | 33.94 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr11_+_86085778 | 33.94 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr13_+_50589390 | 33.84 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr3_-_167098059 | 33.65 |
ENST00000392764.1
ENST00000474464.1 ENST00000392766.2 ENST00000485651.1 |
ZBBX
|
zinc finger, B-box domain containing |
| chr13_+_21141270 | 32.73 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr13_+_21141208 | 31.97 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr1_-_60539405 | 31.81 |
ENST00000450089.2
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr12_-_113658892 | 31.61 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr1_-_60539422 | 31.39 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr10_+_82116529 | 31.35 |
ENST00000411538.1
ENST00000256039.2 |
DYDC2
|
DPY30 domain containing 2 |
| chr6_+_52285131 | 30.70 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr10_-_82116467 | 30.57 |
ENST00000454362.1
|
DYDC1
|
DPY30 domain containing 1 |
| chr3_-_197676740 | 30.39 |
ENST00000452735.1
ENST00000453254.1 ENST00000455191.1 |
IQCG
|
IQ motif containing G |
| chr11_+_113185251 | 30.27 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr2_+_61293021 | 29.68 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr1_-_75139397 | 29.67 |
ENST00000326665.5
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr12_-_71551868 | 29.59 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr11_-_47736896 | 29.50 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr6_+_112408768 | 29.43 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr5_-_110062349 | 29.33 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr6_-_33041378 | 29.22 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr5_-_110062384 | 28.99 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr15_+_71185148 | 28.68 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr15_+_71184931 | 28.50 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr22_-_50970566 | 28.36 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr3_+_39149298 | 28.11 |
ENST00000440121.1
|
TTC21A
|
tetratricopeptide repeat domain 21A |
| chr19_-_55791431 | 28.04 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr11_-_8615507 | 27.95 |
ENST00000431279.2
ENST00000418597.1 |
STK33
|
serine/threonine kinase 33 |
| chr3_+_39149145 | 27.90 |
ENST00000301819.6
ENST00000431162.2 |
TTC21A
|
tetratricopeptide repeat domain 21A |
| chr12_-_71551652 | 27.59 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr10_+_106113515 | 27.53 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr12_-_49582978 | 27.39 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_-_108422926 | 27.10 |
ENST00000428840.1
ENST00000526312.1 |
EXPH5
|
exophilin 5 |
| chr16_-_30773372 | 26.97 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr2_+_106682135 | 26.64 |
ENST00000437659.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr12_-_58329819 | 26.13 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr20_-_18447667 | 25.73 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr2_-_62081254 | 25.46 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr16_-_75590114 | 25.41 |
ENST00000568377.1
ENST00000565067.1 ENST00000258173.6 |
TMEM231
|
transmembrane protein 231 |
| chr2_-_62081148 | 25.32 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr16_-_776431 | 25.09 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr2_-_27851843 | 24.67 |
ENST00000324364.3
|
CCDC121
|
coiled-coil domain containing 121 |
| chr3_+_93698974 | 24.63 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr15_+_67547163 | 24.61 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr12_+_113587558 | 24.60 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr2_+_219536749 | 24.40 |
ENST00000295709.3
ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36
|
serine/threonine kinase 36 |
| chr13_+_24153488 | 24.02 |
ENST00000382258.4
ENST00000382263.3 |
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_219537015 | 23.95 |
ENST00000440309.1
ENST00000424080.1 |
STK36
|
serine/threonine kinase 36 |
| chr4_-_2420335 | 23.90 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr10_-_28287968 | 23.64 |
ENST00000305242.5
|
ARMC4
|
armadillo repeat containing 4 |
| chr2_+_73612858 | 23.62 |
ENST00000409009.1
ENST00000264448.6 ENST00000377715.1 |
ALMS1
|
Alstrom syndrome 1 |
| chr2_+_95537170 | 23.50 |
ENST00000295201.4
|
TEKT4
|
tektin 4 |
| chr5_+_121465207 | 23.20 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr15_+_74610894 | 23.07 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr2_-_63815860 | 23.07 |
ENST00000272321.7
ENST00000431065.1 |
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr14_+_75536335 | 22.99 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr15_-_56757329 | 22.57 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr11_+_537494 | 22.49 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr14_+_75536280 | 22.43 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_+_105452094 | 22.38 |
ENST00000551606.1
ENST00000547315.1 |
C14orf79
|
chromosome 14 open reading frame 79 |
| chr19_-_55791563 | 22.27 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr2_-_207629997 | 22.23 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr15_+_81426588 | 22.23 |
ENST00000286732.4
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr16_-_67450325 | 21.84 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr1_-_118727781 | 21.54 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr11_+_124543694 | 21.38 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr2_-_207630033 | 21.18 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chrX_+_35937843 | 21.17 |
ENST00000297866.5
|
CXorf22
|
chromosome X open reading frame 22 |
| chr6_-_165723088 | 21.08 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr6_+_52285046 | 20.97 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr1_+_118148556 | 20.36 |
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C |
| chr19_-_55791058 | 20.33 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr21_-_47738112 | 20.27 |
ENST00000417060.1
|
C21orf58
|
chromosome 21 open reading frame 58 |
| chr1_+_20512568 | 20.23 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr7_+_158649242 | 19.69 |
ENST00000407559.3
|
WDR60
|
WD repeat domain 60 |
| chr2_-_230579185 | 19.53 |
ENST00000341772.4
|
DNER
|
delta/notch-like EGF repeat containing |
| chr11_+_113185292 | 19.49 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr19_-_55791540 | 19.36 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_+_55996565 | 19.32 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr19_+_55996316 | 19.27 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr6_+_151815143 | 19.26 |
ENST00000239374.7
ENST00000367290.5 |
CCDC170
|
coiled-coil domain containing 170 |
| chr6_-_43478239 | 19.26 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr7_+_89874524 | 19.12 |
ENST00000497910.1
|
C7orf63
|
chromosome 7 open reading frame 63 |
| chr12_+_122667658 | 18.87 |
ENST00000339777.4
ENST00000425921.1 |
LRRC43
|
leucine rich repeat containing 43 |
| chr7_+_89874483 | 18.85 |
ENST00000389297.4
ENST00000316089.8 |
C7orf63
|
chromosome 7 open reading frame 63 |
| chr12_+_49297887 | 18.64 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr8_+_144798429 | 18.59 |
ENST00000338033.4
ENST00000395107.4 ENST00000395108.2 |
MAPK15
|
mitogen-activated protein kinase 15 |
| chr2_+_219187871 | 18.41 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr2_+_39103103 | 18.38 |
ENST00000340556.6
ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2
|
MORN repeat containing 2 |
| chr4_+_15471489 | 18.30 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr11_-_8615720 | 18.14 |
ENST00000358872.3
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr6_+_146920097 | 18.10 |
ENST00000397944.3
ENST00000522242.1 |
ADGB
|
androglobin |
| chr1_+_1109272 | 18.09 |
ENST00000379290.1
ENST00000379289.1 |
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr6_-_159421198 | 18.05 |
ENST00000252655.1
ENST00000297262.3 ENST00000367069.2 |
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr2_+_170550944 | 18.04 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr11_-_8615488 | 17.95 |
ENST00000315204.1
ENST00000396672.1 ENST00000396673.1 |
STK33
|
serine/threonine kinase 33 |
| chr3_-_167371740 | 17.76 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr11_+_113185533 | 17.59 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr3_-_112565703 | 17.30 |
ENST00000488794.1
|
CD200R1L
|
CD200 receptor 1-like |
| chr6_+_146920116 | 17.19 |
ENST00000367493.3
|
ADGB
|
androglobin |
| chr17_-_56296580 | 17.07 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr11_+_6260298 | 16.91 |
ENST00000379936.2
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr4_-_156298028 | 16.90 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr7_+_138818490 | 16.73 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr9_-_90589586 | 16.53 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr17_+_73642486 | 16.49 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr2_+_26785409 | 16.26 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr1_+_245133062 | 16.21 |
ENST00000366523.1
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr6_-_159420780 | 16.01 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr14_+_74111578 | 15.90 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chr4_+_81256871 | 15.83 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr1_+_32674675 | 15.82 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr9_-_90589402 | 15.76 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr21_-_35883541 | 15.49 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr17_+_73642315 | 15.48 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr4_-_156298087 | 15.42 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr3_-_47324079 | 15.26 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr6_+_147091575 | 15.07 |
ENST00000326916.8
ENST00000470716.2 ENST00000367488.1 |
ADGB
|
androglobin |
| chr11_+_36616355 | 15.05 |
ENST00000532470.2
|
C11orf74
|
chromosome 11 open reading frame 74 |
| chr3_+_158288942 | 15.05 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr18_-_47792851 | 14.83 |
ENST00000398545.4
|
CCDC11
|
coiled-coil domain containing 11 |
| chr3_+_158288960 | 14.72 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 42.4 | 169.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 21.9 | 87.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 19.5 | 78.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 18.4 | 129.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 17.3 | 103.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 14.7 | 308.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 12.5 | 37.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 11.3 | 383.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 10.9 | 153.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 10.1 | 141.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 8.2 | 24.6 | GO:0061565 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 6.6 | 132.1 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 6.5 | 19.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 6.3 | 113.8 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 6.1 | 18.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 6.0 | 35.9 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) glomerular visceral epithelial cell migration(GO:0090521) |
| 5.8 | 5.8 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 5.5 | 364.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 5.2 | 46.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 4.5 | 18.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 4.4 | 13.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 4.3 | 21.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 4.0 | 75.7 | GO:0003341 | cilium movement(GO:0003341) |
| 3.8 | 26.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 3.4 | 17.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 3.4 | 16.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 3.1 | 43.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 3.1 | 9.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 3.0 | 6.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 2.9 | 49.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 2.9 | 46.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 2.6 | 20.9 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 2.6 | 23.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 2.3 | 6.9 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 2.2 | 8.9 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 2.0 | 4.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 1.8 | 12.8 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 1.8 | 17.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 1.7 | 5.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.7 | 5.0 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 1.6 | 207.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 1.6 | 4.8 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.6 | 26.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.5 | 9.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.5 | 5.9 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.5 | 4.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.4 | 54.6 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 1.3 | 38.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 1.3 | 1.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 1.3 | 72.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 1.2 | 27.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.2 | 6.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.2 | 14.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 1.2 | 23.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.1 | 9.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 1.1 | 2.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 1.1 | 4.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.0 | 3.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 1.0 | 2.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.9 | 2.8 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.9 | 5.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.9 | 12.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.9 | 2.7 | GO:0046533 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) |
| 0.9 | 6.9 | GO:0070092 | negative regulation of glycogen biosynthetic process(GO:0045719) regulation of glucagon secretion(GO:0070092) |
| 0.9 | 197.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.8 | 5.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.8 | 10.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.8 | 3.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.8 | 12.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.8 | 3.2 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.8 | 7.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.8 | 18.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.8 | 12.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.7 | 2.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.7 | 8.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.7 | 2.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.7 | 49.1 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.7 | 2.2 | GO:2001162 | histone H3-K79 methylation(GO:0034729) regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.7 | 2.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 82.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.7 | 97.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.7 | 243.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.7 | 19.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.7 | 2.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.6 | 3.2 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.6 | 6.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.6 | 1.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.6 | 3.0 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 9.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.6 | 47.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.6 | 4.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.6 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.6 | 2.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.5 | 5.5 | GO:1902116 | negative regulation of organelle assembly(GO:1902116) |
| 0.5 | 12.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 1.0 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.5 | 12.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.5 | 2.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 1.8 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.4 | 2.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.4 | 3.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 1.3 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 2.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 6.1 | GO:0032990 | cell part morphogenesis(GO:0032990) |
| 0.4 | 8.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 9.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.4 | 2.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.4 | 2.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 1.7 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.3 | 2.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 2.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 1.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 2.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.3 | 1.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.3 | 61.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.3 | 27.9 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.3 | 5.7 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.3 | 6.9 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.3 | 2.6 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 44.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.3 | 37.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.3 | 8.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.3 | 4.7 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.3 | 2.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 7.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 1.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.3 | 1.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 2.0 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 1.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 2.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 1.2 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.2 | 1.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 8.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 3.0 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.2 | 0.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 14.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 0.8 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 1.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 4.9 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 1.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 4.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.2 | 14.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 7.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 3.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 2.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 5.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 7.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 15.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 0.5 | GO:0010836 | base-excision repair, DNA ligation(GO:0006288) negative regulation of protein ADP-ribosylation(GO:0010836) regulation of single strand break repair(GO:1903516) |
| 0.1 | 4.0 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 10.4 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.1 | 1.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.7 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 5.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 52.8 | GO:0031344 | regulation of cell projection organization(GO:0031344) |
| 0.1 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 5.1 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.1 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.3 | GO:1903299 | regulation of glucokinase activity(GO:0033131) regulation of hexokinase activity(GO:1903299) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 1.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.3 | GO:0032940 | secretion by cell(GO:0032940) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.3 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 1.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.5 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.4 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 51.3 | 256.7 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 25.5 | 280.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 20.3 | 60.8 | GO:0001534 | radial spoke(GO:0001534) |
| 17.4 | 121.5 | GO:0002177 | manchette(GO:0002177) |
| 15.6 | 296.5 | GO:0036038 | MKS complex(GO:0036038) |
| 15.2 | 75.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 14.2 | 170.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 7.8 | 23.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 7.7 | 161.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 6.7 | 6.7 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 6.6 | 19.7 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 6.3 | 182.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 5.6 | 56.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 5.4 | 113.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 5.4 | 26.8 | GO:0098536 | deuterosome(GO:0098536) |
| 5.3 | 94.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 5.2 | 51.8 | GO:0044447 | axoneme part(GO:0044447) |
| 4.5 | 18.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 4.4 | 377.3 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 4.1 | 32.7 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 3.3 | 87.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 3.0 | 168.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 2.8 | 46.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 2.7 | 24.1 | GO:0034464 | BBSome(GO:0034464) |
| 2.6 | 139.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 2.2 | 94.5 | GO:0030286 | dynein complex(GO:0030286) |
| 1.7 | 70.1 | GO:0031514 | motile cilium(GO:0031514) |
| 1.4 | 101.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 1.3 | 80.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 1.1 | 8.0 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.9 | 25.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.9 | 7.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 52.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.8 | 20.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.8 | 12.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.8 | 2.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.7 | 18.4 | GO:0097223 | sperm part(GO:0097223) |
| 0.7 | 8.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.7 | 186.9 | GO:0005929 | cilium(GO:0005929) |
| 0.6 | 5.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 3.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.6 | 2.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 2.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.5 | 19.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 2.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 2.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 1.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.4 | 2.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.4 | 5.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 5.0 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.4 | 2.9 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.3 | 7.6 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.3 | 23.0 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 2.2 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 3.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 1.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 3.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 2.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 2.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 1.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 6.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 2.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 9.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 9.2 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 1.6 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.2 | 8.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 81.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 28.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 21.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 8.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 5.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 7.0 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 2.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 8.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 11.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.5 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 4.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.7 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 7.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.6 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 8.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 53.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 2.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 4.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 4.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 19.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 132.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 13.2 | 289.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 10.8 | 32.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 8.5 | 59.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 7.5 | 104.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 7.3 | 72.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 6.7 | 188.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 6.7 | 161.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 6.2 | 43.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 6.1 | 18.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 4.6 | 113.8 | GO:0031005 | filamin binding(GO:0031005) |
| 4.3 | 12.9 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 4.1 | 12.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 4.0 | 95.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 3.7 | 190.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 3.2 | 258.9 | GO:0043621 | protein self-association(GO:0043621) |
| 3.0 | 18.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.9 | 17.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 2.4 | 12.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.8 | 9.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.7 | 5.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.6 | 4.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.5 | 15.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.4 | 4.3 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 1.4 | 8.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.4 | 6.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 1.2 | 20.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 1.2 | 4.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.2 | 8.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.1 | 6.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.1 | 6.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 1.0 | 5.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.9 | 38.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.8 | 37.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.8 | 18.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.8 | 59.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.7 | 9.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.7 | 31.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 2.2 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.7 | 10.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 19.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 3.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.6 | 24.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.6 | 2.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 2.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.6 | 2.2 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.5 | 2.7 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.5 | 2.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 2.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.5 | 2.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 8.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 23.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.5 | 19.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.5 | 12.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 16.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 8.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.4 | 23.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 82.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.4 | 2.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) beta-glucosidase activity(GO:0008422) |
| 0.4 | 1.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.4 | 1.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.4 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 92.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 201.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.4 | 4.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 2.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 9.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 6.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 2.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 8.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 10.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 1.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 16.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 5.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 10.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 10.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 2.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 9.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 3.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 5.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 2.6 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 4.0 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.2 | 24.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.2 | 10.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 4.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 3.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 2.8 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 11.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 26.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.2 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 14.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 2.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 21.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 9.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 11.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 59.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.5 | GO:0032356 | oxidized DNA binding(GO:0032356) |
| 0.1 | 8.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 6.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 29.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 1.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 4.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 15.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 116.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 1.4 | 69.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.8 | 7.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.6 | 38.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.5 | 6.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 25.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 6.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 18.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 8.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 6.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 8.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 4.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 38.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 7.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 38.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.4 | 52.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.3 | 29.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 12.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 33.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 41.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 24.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.4 | 9.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 12.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 4.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 5.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 2.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 5.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 6.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.1 | 2.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 6.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 6.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 5.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |