Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for RORC
Z-value: 0.40
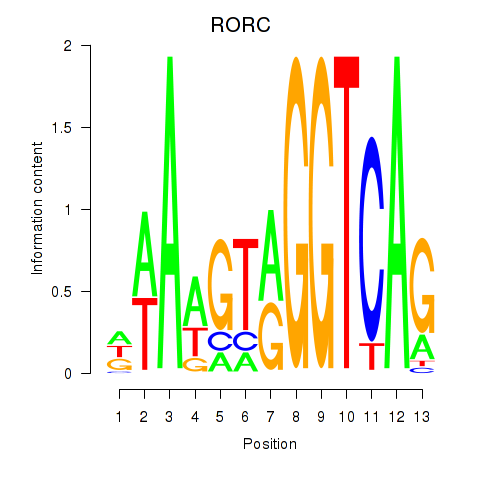
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORC | hg19_v2_chr1_-_151798546_151798590 | 0.31 | 9.4e-02 | Click! |
Activity profile of RORC motif
Sorted Z-values of RORC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4671352 | 0.75 |
ENST00000542744.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr12_+_7167980 | 0.62 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr1_+_104198377 | 0.50 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr8_+_71485681 | 0.46 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr3_-_45957088 | 0.42 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr17_-_46623441 | 0.40 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr3_-_45957534 | 0.35 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chrX_-_102531717 | 0.34 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr6_-_46922659 | 0.29 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr7_-_128415844 | 0.29 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr19_+_38924316 | 0.29 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr3_+_160559931 | 0.29 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr10_-_21435488 | 0.27 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr19_-_10450287 | 0.24 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_-_1821496 | 0.23 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_-_49140609 | 0.22 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chrX_-_43832711 | 0.21 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr12_+_56414795 | 0.20 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_+_93963590 | 0.20 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr9_-_110251836 | 0.18 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr19_-_49140692 | 0.18 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_+_93964158 | 0.17 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr3_-_65583561 | 0.17 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_-_240906911 | 0.17 |
ENST00000431139.2
|
RP11-80B9.4
|
RP11-80B9.4 |
| chr19_+_1269324 | 0.15 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr14_-_23451845 | 0.15 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr20_-_23433494 | 0.14 |
ENST00000377007.3
|
CST11
|
cystatin 11 |
| chr16_-_69385681 | 0.13 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr4_+_85504075 | 0.12 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr20_+_37209820 | 0.10 |
ENST00000537425.1
ENST00000373348.3 ENST00000416116.1 |
ADIG
|
adipogenin |
| chr12_-_39734783 | 0.10 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr12_-_54652060 | 0.10 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr7_-_43965937 | 0.09 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr2_+_148778570 | 0.09 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr12_-_82152420 | 0.09 |
ENST00000552948.1
ENST00000548586.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_-_217311090 | 0.08 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr7_+_20687017 | 0.08 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_+_26869597 | 0.08 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr14_+_21785693 | 0.08 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr1_-_185286461 | 0.07 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr20_-_33732952 | 0.07 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr16_+_1832902 | 0.07 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr7_-_141541221 | 0.07 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr19_+_12780512 | 0.06 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr4_+_140586922 | 0.06 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr2_-_148778323 | 0.06 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr10_-_121296045 | 0.06 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr17_-_48785216 | 0.06 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_+_65292884 | 0.06 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr1_-_153522562 | 0.06 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_114447763 | 0.06 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr7_+_112063192 | 0.06 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr14_-_23451467 | 0.05 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr17_-_37764128 | 0.05 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr2_+_160590469 | 0.05 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr16_+_24266874 | 0.05 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr1_-_145826450 | 0.04 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr19_-_8567478 | 0.04 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr3_+_185303962 | 0.04 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr3_-_42452050 | 0.03 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chr18_-_23671139 | 0.03 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr2_+_119699864 | 0.03 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr1_-_62190793 | 0.03 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr6_+_43738444 | 0.03 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr10_-_103578162 | 0.02 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr19_-_39735646 | 0.02 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr6_+_44094627 | 0.02 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr8_-_95449155 | 0.01 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr12_+_56624436 | 0.01 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr7_+_20686946 | 0.01 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_+_119699742 | 0.01 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr5_+_177557997 | 0.00 |
ENST00000313386.4
ENST00000515098.1 ENST00000542098.1 ENST00000502814.1 ENST00000507457.1 ENST00000508647.1 |
RMND5B
|
required for meiotic nuclear division 5 homolog B (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |