Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
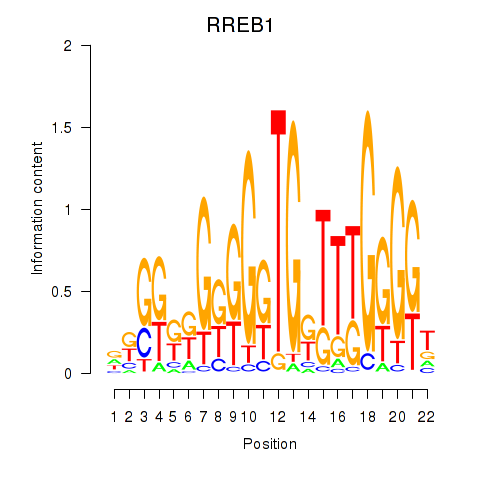
Results for RREB1
Z-value: 1.29
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107830_7107993 | 0.27 | 1.5e-01 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51471362 | 8.62 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr19_-_51471381 | 8.47 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr19_-_51456198 | 6.39 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 6.34 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr19_-_51456321 | 6.34 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr6_+_151646800 | 4.67 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr18_-_21242774 | 4.45 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr4_+_75230853 | 3.90 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr12_+_96588279 | 3.81 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr2_+_234104079 | 3.37 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_-_19051927 | 3.35 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr18_-_21242833 | 3.28 |
ENST00000586087.1
ENST00000592179.1 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr19_-_19051993 | 2.97 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr16_-_4588762 | 2.92 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr1_-_17307173 | 2.75 |
ENST00000438542.1
ENST00000375535.3 |
MFAP2
|
microfibrillar-associated protein 2 |
| chr10_+_17271266 | 2.74 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr16_-_4588822 | 2.73 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr5_+_150591678 | 2.60 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chr9_+_131174024 | 2.44 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr16_+_447226 | 2.36 |
ENST00000433358.1
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr16_+_447209 | 2.34 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr16_+_58533951 | 2.27 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr11_+_69455855 | 2.23 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chrX_+_17755563 | 2.13 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr16_-_4588469 | 2.13 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr10_-_121356518 | 2.05 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr17_+_1958388 | 2.03 |
ENST00000399849.3
|
HIC1
|
hypermethylated in cancer 1 |
| chr2_-_85637459 | 1.99 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_+_192543153 | 1.98 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr7_-_24797032 | 1.96 |
ENST00000409970.1
ENST00000409775.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr18_+_33877654 | 1.92 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr8_-_144655141 | 1.90 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr2_+_28618532 | 1.79 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr10_-_71930222 | 1.74 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr17_-_39637392 | 1.73 |
ENST00000246639.2
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chr19_-_39226045 | 1.69 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr17_-_39216344 | 1.66 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr20_+_58179582 | 1.60 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_-_102257139 | 1.54 |
ENST00000521076.1
ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4
|
RAS p21 protein activator 4 |
| chr1_-_228604328 | 1.48 |
ENST00000355586.4
ENST00000366698.2 ENST00000520264.1 ENST00000479800.1 ENST00000295033.3 |
TRIM17
|
tripartite motif containing 17 |
| chr8_+_32405785 | 1.47 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr1_-_6240183 | 1.47 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr17_-_73840415 | 1.42 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr7_-_128045984 | 1.41 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr21_-_47648665 | 1.39 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr15_-_70390191 | 1.38 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr15_+_21145765 | 1.37 |
ENST00000553416.1
|
CT60
|
cancer/testis antigen 60 (non-protein coding) |
| chrX_+_134166333 | 1.35 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr12_-_8088871 | 1.31 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr17_-_31620006 | 1.30 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr4_+_123747979 | 1.28 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr2_+_192542850 | 1.25 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr15_+_63481668 | 1.20 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr8_-_142011036 | 1.19 |
ENST00000520892.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_228337079 | 1.18 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr20_-_36793663 | 1.17 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr14_+_100070869 | 1.17 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr3_+_154797877 | 1.15 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr20_+_23168759 | 1.15 |
ENST00000411595.1
|
RP4-737E23.2
|
RP4-737E23.2 |
| chr17_+_58499844 | 1.14 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr9_+_35906176 | 1.12 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chrX_-_119249819 | 1.12 |
ENST00000217999.2
|
RHOXF1
|
Rhox homeobox family, member 1 |
| chr1_-_161193349 | 1.12 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr22_-_36357671 | 1.11 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_56415100 | 1.09 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr3_-_42744312 | 1.05 |
ENST00000416756.1
ENST00000441594.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr15_-_89764929 | 1.03 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_+_110091189 | 1.03 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr12_-_53074182 | 1.02 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr9_-_21994597 | 1.00 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr5_-_151304337 | 0.97 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr14_+_105155925 | 0.97 |
ENST00000330634.7
ENST00000398337.4 ENST00000392634.4 |
INF2
|
inverted formin, FH2 and WH2 domain containing |
| chr12_-_49351148 | 0.96 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr12_-_120687948 | 0.95 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr8_+_32406179 | 0.95 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chrX_-_153881842 | 0.94 |
ENST00000369585.3
ENST00000247306.4 |
CTAG2
|
cancer/testis antigen 2 |
| chr20_+_34700333 | 0.94 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_+_151903207 | 0.92 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1 |
| chr16_+_27325202 | 0.92 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chrX_+_47082408 | 0.91 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chrX_+_153813407 | 0.91 |
ENST00000443287.2
ENST00000333128.3 |
CTAG1A
|
cancer/testis antigen 1A |
| chr12_-_95044309 | 0.91 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_+_28889801 | 0.91 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr4_-_103940791 | 0.91 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr4_+_123747834 | 0.91 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr3_-_116163830 | 0.90 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr19_-_18654293 | 0.89 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr11_-_59436453 | 0.87 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr19_-_51336443 | 0.86 |
ENST00000598673.1
|
KLK15
|
kallikrein-related peptidase 15 |
| chr3_-_39196049 | 0.86 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chrX_+_151903253 | 0.86 |
ENST00000452779.2
ENST00000370291.2 |
CSAG1
|
chondrosarcoma associated gene 1 |
| chr19_+_40973049 | 0.85 |
ENST00000598249.1
ENST00000338932.3 ENST00000344104.3 |
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr21_-_36259445 | 0.84 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr22_-_51066521 | 0.81 |
ENST00000395621.3
ENST00000395619.3 ENST00000356098.5 ENST00000216124.5 ENST00000453344.2 ENST00000547307.1 ENST00000547805.1 |
ARSA
|
arylsulfatase A |
| chr19_-_18653781 | 0.80 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr22_-_37976082 | 0.80 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr1_-_153363452 | 0.78 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr16_+_28889703 | 0.78 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_-_39222131 | 0.77 |
ENST00000394015.2
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr2_-_101034070 | 0.76 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr4_+_190992087 | 0.76 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7 |
| chr3_+_186435137 | 0.76 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr4_+_191005567 | 0.75 |
ENST00000605758.1
ENST00000554285.2 |
DUX4
|
double homeobox 4 |
| chr4_+_191012160 | 0.75 |
ENST00000553702.1
ENST00000554376.1 |
DUX4L2
|
double homeobox 4 like 2 |
| chr4_+_190998673 | 0.75 |
ENST00000554668.1
|
DUX4L5
|
double homeobox 4 like 5 |
| chr4_+_191005267 | 0.75 |
ENST00000556625.1
|
DUX4
|
double homeobox 4 |
| chr4_+_191011860 | 0.75 |
ENST00000440426.1
|
DUX4L2
|
double homeobox 4 like 2 |
| chr4_+_147560042 | 0.75 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2 |
| chr4_+_190998973 | 0.75 |
ENST00000605696.1
ENST00000557283.2 |
DUX4L5
|
double homeobox 4 like 5 |
| chr4_+_191008560 | 0.75 |
ENST00000554690.1
|
DUX4L3
|
double homeobox 4 like 3 |
| chr12_-_12715266 | 0.74 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chrX_-_152939252 | 0.74 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr19_+_55477711 | 0.74 |
ENST00000448584.2
ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr8_+_32406137 | 0.74 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr16_+_28834531 | 0.74 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like |
| chr19_+_55476620 | 0.74 |
ENST00000543010.1
ENST00000391721.4 ENST00000339757.7 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr3_-_48471454 | 0.73 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr9_-_21994344 | 0.73 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr9_-_34126730 | 0.73 |
ENST00000361264.4
|
DCAF12
|
DDB1 and CUL4 associated factor 12 |
| chr11_-_71791435 | 0.72 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr4_+_191008860 | 0.72 |
ENST00000605191.1
ENST00000557656.2 |
DUX4L3
|
double homeobox 4 like 3 |
| chr4_+_190995680 | 0.72 |
ENST00000605381.1
ENST00000553722.2 |
DUX4L6
|
double homeobox 4 like 6 |
| chr4_+_190995380 | 0.72 |
ENST00000555191.1
|
DUX4L6
|
double homeobox 4 like 6 |
| chrX_-_153847522 | 0.71 |
ENST00000328435.2
ENST00000359887.4 |
CTAG1B
|
cancer/testis antigen 1B |
| chr19_-_51222707 | 0.71 |
ENST00000391814.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr17_-_73840774 | 0.71 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr7_-_92465868 | 0.70 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr19_-_15560730 | 0.70 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr14_+_23563952 | 0.69 |
ENST00000319074.4
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chrX_+_151867214 | 0.69 |
ENST00000329342.5
ENST00000412733.1 ENST00000457643.1 |
MAGEA6
|
melanoma antigen family A, 6 |
| chrX_-_13752675 | 0.69 |
ENST00000380579.1
ENST00000458511.2 ENST00000519885.1 ENST00000358231.5 ENST00000518847.1 ENST00000453655.2 ENST00000359680.5 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr7_-_752074 | 0.68 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr12_+_118454500 | 0.68 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr10_-_103603523 | 0.68 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr17_-_77005860 | 0.67 |
ENST00000591773.1
ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr8_-_60031762 | 0.67 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr17_-_62658186 | 0.67 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr11_-_85779971 | 0.67 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr17_+_15604513 | 0.66 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chrX_-_131352152 | 0.66 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr11_+_46402744 | 0.65 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_+_53078273 | 0.65 |
ENST00000332582.4
|
GPR173
|
G protein-coupled receptor 173 |
| chr2_-_32236002 | 0.65 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr12_+_96588143 | 0.64 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr11_-_71791518 | 0.63 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr8_-_22550815 | 0.63 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr12_-_129570545 | 0.63 |
ENST00000389441.4
|
TMEM132D
|
transmembrane protein 132D |
| chr1_-_155947951 | 0.63 |
ENST00000313695.7
ENST00000497907.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr13_-_52027134 | 0.62 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_+_75498739 | 0.61 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr12_+_119419294 | 0.61 |
ENST00000267260.4
|
SRRM4
|
serine/arginine repetitive matrix 4 |
| chr17_-_7082861 | 0.60 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr8_+_38261880 | 0.60 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr8_+_144816303 | 0.59 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr19_-_8675559 | 0.59 |
ENST00000597188.1
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr17_+_38474489 | 0.59 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr19_-_51192661 | 0.58 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr12_+_62654119 | 0.58 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr12_+_6554021 | 0.58 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr8_-_66753682 | 0.58 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chr2_-_163695128 | 0.58 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr8_-_145331153 | 0.57 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr10_-_75415825 | 0.57 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr12_+_26348582 | 0.57 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr17_+_58499066 | 0.56 |
ENST00000474834.1
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr4_+_190992387 | 0.56 |
ENST00000554906.2
ENST00000553820.2 |
DUX4L7
|
double homeobox 4 like 7 |
| chr19_-_57352064 | 0.56 |
ENST00000326441.9
ENST00000593695.1 ENST00000599577.1 ENST00000594389.1 ENST00000423103.2 ENST00000598410.1 ENST00000593711.1 ENST00000391708.3 ENST00000221722.5 ENST00000599935.1 |
PEG3
ZIM2
|
paternally expressed 3 zinc finger, imprinted 2 |
| chr3_+_154797428 | 0.55 |
ENST00000460393.1
|
MME
|
membrane metallo-endopeptidase |
| chr1_-_31712401 | 0.54 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr16_+_21169976 | 0.54 |
ENST00000572258.1
ENST00000261388.3 ENST00000451578.2 ENST00000572599.1 ENST00000577162.1 |
TMEM159
|
transmembrane protein 159 |
| chr3_+_8775466 | 0.54 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr4_-_6474173 | 0.54 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr17_-_58499698 | 0.53 |
ENST00000590297.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr11_+_128562372 | 0.52 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_2323089 | 0.50 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chrX_-_151307020 | 0.50 |
ENST00000370323.4
ENST00000244096.3 ENST00000427322.2 |
MAGEA10
|
melanoma antigen family A, 10 |
| chr11_-_64703354 | 0.49 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr2_+_127066059 | 0.49 |
ENST00000435352.1
|
AC023347.1
|
AC023347.1 |
| chr4_+_56719782 | 0.49 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr19_+_35609380 | 0.49 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr12_+_54447637 | 0.49 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr5_+_151151471 | 0.49 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr17_-_4167142 | 0.48 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr11_-_85780086 | 0.48 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_-_57940904 | 0.48 |
ENST00000550954.1
ENST00000434715.3 ENST00000546670.1 ENST00000543672.1 |
DCTN2
|
dynactin 2 (p50) |
| chr8_+_73449625 | 0.48 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr15_-_70390213 | 0.48 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr8_+_145064233 | 0.48 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr2_-_202508169 | 0.47 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chr11_+_46403194 | 0.47 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr12_-_57941004 | 0.46 |
ENST00000550750.1
ENST00000548249.1 |
DCTN2
|
dynactin 2 (p50) |
| chrX_-_153979315 | 0.46 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr10_-_103603677 | 0.46 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr16_+_68771128 | 0.46 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr11_+_46403303 | 0.45 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr20_+_33462888 | 0.44 |
ENST00000336325.4
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr18_+_55102917 | 0.44 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr14_+_74353320 | 0.44 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr12_+_44152740 | 0.44 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr1_-_28559502 | 0.43 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.3 | 3.9 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 1.1 | 3.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.8 | 17.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.6 | 1.7 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 1.7 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.5 | 3.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.4 | 1.3 | GO:0050894 | determination of affect(GO:0050894) |
| 0.4 | 1.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 6.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.4 | 1.1 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.4 | 3.9 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.3 | 1.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 1.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 2.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.3 | 4.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 2.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 2.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 2.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.6 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 0.8 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.2 | 0.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 6.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 2.7 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 1.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 2.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 1.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.9 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.2 | 0.7 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.2 | 1.2 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.7 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 2.1 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.6 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.4 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.5 | GO:0033292 | T-tubule organization(GO:0033292) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 2.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 2.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 1.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 7.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 2.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.5 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.2 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:1904847 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.4 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 1.0 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.3 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 3.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 4.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 2.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) cranial suture morphogenesis(GO:0060363) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 1.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 19.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.0 | 7.8 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 2.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.6 | 3.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 1.7 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.2 | 0.7 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.2 | 1.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 3.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 2.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 6.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 2.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 8.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 3.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 4.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 15.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 3.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.5 | 4.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 1.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.4 | 1.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.4 | 6.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 2.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 3.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 0.9 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.3 | 1.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 4.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 0.7 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 0.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 1.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.5 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 1.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.1 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.1 | 2.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 33.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 4.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 0.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.4 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 1.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 2.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 2.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 1.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 5.4 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 5.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 2.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 4.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |