Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for SIX1_SIX3_SIX2
Z-value: 0.54
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
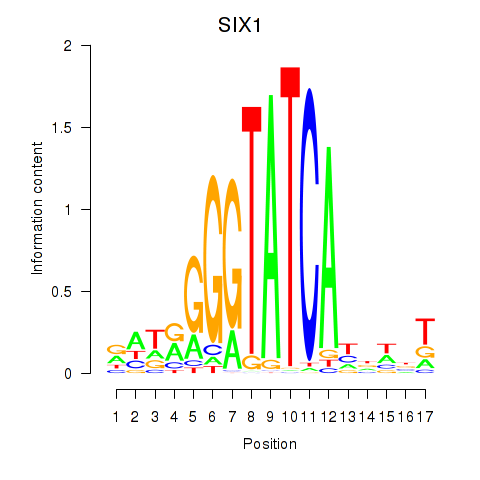
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
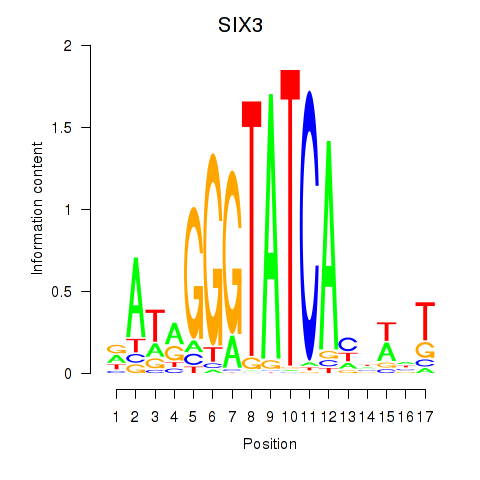
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
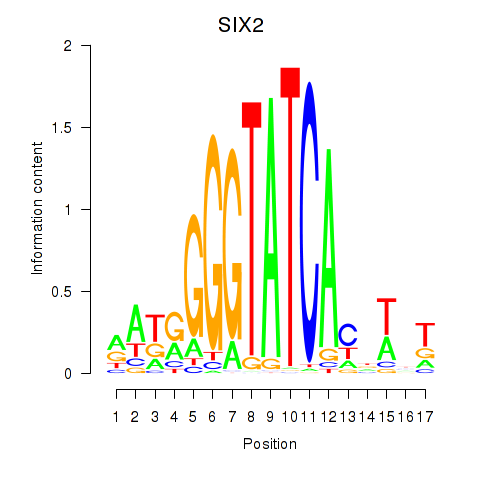
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.27 | 1.4e-01 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | -0.08 | 6.7e-01 | Click! |
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.05 | 8.0e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_58499434 | 0.86 |
ENST00000344743.3
ENST00000278400.3 |
GLYAT
|
glycine-N-acyltransferase |
| chr11_-_32452357 | 0.83 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr1_-_13115578 | 0.78 |
ENST00000414205.2
|
PRAMEF6
|
PRAME family member 6 |
| chr20_-_55100981 | 0.78 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr13_-_46679185 | 0.74 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 0.74 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_+_26737253 | 0.73 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr1_+_26737292 | 0.71 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr9_+_12693336 | 0.70 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr1_-_13117736 | 0.69 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr1_+_13359819 | 0.65 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr7_+_123241908 | 0.61 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chrX_-_65259914 | 0.59 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_65259900 | 0.59 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_240906911 | 0.59 |
ENST00000431139.2
|
RP11-80B9.4
|
RP11-80B9.4 |
| chrX_+_101470280 | 0.52 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr1_+_182419261 | 0.51 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr3_+_118866222 | 0.50 |
ENST00000490594.1
|
RP11-484M3.5
|
Uncharacterized protein |
| chr1_+_241695424 | 0.49 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_+_41594377 | 0.46 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr21_+_27011899 | 0.45 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr7_-_14880892 | 0.43 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr10_-_134331695 | 0.43 |
ENST00000455414.1
|
RP11-432J24.5
|
RP11-432J24.5 |
| chr2_-_207629997 | 0.40 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr2_-_207630033 | 0.39 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr15_+_40650408 | 0.38 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr19_-_58446721 | 0.38 |
ENST00000396147.1
ENST00000595569.1 ENST00000599852.1 ENST00000425570.3 ENST00000601593.1 |
ZNF418
|
zinc finger protein 418 |
| chr6_-_76203454 | 0.38 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_-_12946025 | 0.38 |
ENST00000235349.5
|
PRAMEF4
|
PRAME family member 4 |
| chr8_+_118147498 | 0.36 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr14_+_22739823 | 0.36 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr3_+_149191723 | 0.36 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_-_13007420 | 0.35 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr3_-_46506358 | 0.32 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr11_+_73661364 | 0.32 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr1_-_27998689 | 0.32 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr1_+_241695670 | 0.31 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_-_103603523 | 0.31 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_+_51236703 | 0.31 |
ENST00000551456.1
ENST00000398458.3 |
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12 |
| chr2_-_105953912 | 0.31 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr18_+_61575200 | 0.30 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr1_-_44818599 | 0.29 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr14_+_62462541 | 0.29 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr1_-_108743471 | 0.28 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr1_+_160709029 | 0.28 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr17_-_3030875 | 0.27 |
ENST00000328890.2
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1 |
| chr11_+_71791359 | 0.26 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr3_+_42850959 | 0.26 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr8_-_134115118 | 0.26 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr1_-_60539422 | 0.26 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr11_-_35547151 | 0.25 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr7_+_138915102 | 0.25 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr17_-_55911970 | 0.25 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr10_+_7745232 | 0.24 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr19_+_11457175 | 0.24 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr16_+_21695750 | 0.23 |
ENST00000388956.4
|
OTOA
|
otoancorin |
| chr10_-_103603568 | 0.23 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr1_+_160709055 | 0.23 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr2_-_209010874 | 0.23 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr14_-_24911448 | 0.23 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr19_-_54872556 | 0.23 |
ENST00000444687.1
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr4_+_70894130 | 0.22 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr3_+_111718036 | 0.22 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr11_-_5364809 | 0.21 |
ENST00000300773.2
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5 |
| chr5_+_161112754 | 0.21 |
ENST00000523217.1
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr1_+_196743912 | 0.21 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr10_+_7745303 | 0.20 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_160709076 | 0.20 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr3_+_111718173 | 0.20 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr19_+_11457232 | 0.20 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr18_-_6929797 | 0.20 |
ENST00000581725.1
ENST00000583316.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr7_-_44198874 | 0.20 |
ENST00000395796.3
ENST00000345378.2 |
GCK
|
glucokinase (hexokinase 4) |
| chr3_-_19975665 | 0.20 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr21_-_34185944 | 0.20 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr8_+_99129513 | 0.19 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr10_-_103603677 | 0.19 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr1_+_35225339 | 0.19 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr1_+_40942887 | 0.19 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr3_+_26735991 | 0.19 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr1_-_183387723 | 0.19 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr4_+_80748619 | 0.19 |
ENST00000504263.1
|
PCAT4
|
prostate cancer associated transcript 4 (non-protein coding) |
| chr6_+_31554636 | 0.18 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr1_-_207119738 | 0.18 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr3_+_111717600 | 0.18 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr8_+_76452097 | 0.18 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_157670528 | 0.18 |
ENST00000368186.5
ENST00000496769.1 |
FCRL3
|
Fc receptor-like 3 |
| chr1_+_78769549 | 0.18 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr21_-_35883613 | 0.18 |
ENST00000337385.3
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chrX_-_72095808 | 0.17 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
| chr1_-_169337176 | 0.17 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr12_+_20848486 | 0.17 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_20848377 | 0.17 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr1_+_15256230 | 0.16 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr6_-_167797887 | 0.16 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr14_-_95942173 | 0.16 |
ENST00000334258.5
ENST00000557275.1 ENST00000553340.1 |
SYNE3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr4_+_89299994 | 0.16 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_+_56459198 | 0.16 |
ENST00000291971.3
ENST00000590542.1 |
NLRP8
|
NLR family, pyrin domain containing 8 |
| chr17_-_8151353 | 0.16 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr19_-_55549624 | 0.15 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr21_+_27011584 | 0.15 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr19_-_56249740 | 0.15 |
ENST00000590200.1
ENST00000332836.2 |
NLRP9
|
NLR family, pyrin domain containing 9 |
| chr1_+_207277590 | 0.15 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr19_+_55476620 | 0.15 |
ENST00000543010.1
ENST00000391721.4 ENST00000339757.7 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr6_+_31554779 | 0.15 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr5_-_132073210 | 0.14 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr2_-_220034745 | 0.14 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr11_+_64323428 | 0.14 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr12_+_49121213 | 0.13 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr8_-_95487272 | 0.13 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr6_+_88757507 | 0.13 |
ENST00000237201.1
|
SPACA1
|
sperm acrosome associated 1 |
| chr1_+_6684918 | 0.13 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr6_-_32160622 | 0.13 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr12_-_57882577 | 0.13 |
ENST00000393797.2
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr17_-_72527605 | 0.13 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr17_+_6659153 | 0.13 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr1_-_159046617 | 0.13 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr1_-_208084729 | 0.13 |
ENST00000310833.7
ENST00000356522.4 |
CD34
|
CD34 molecule |
| chr2_-_25391507 | 0.12 |
ENST00000380794.1
|
POMC
|
proopiomelanocortin |
| chr3_-_10452359 | 0.12 |
ENST00000452124.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr22_+_22936998 | 0.12 |
ENST00000390303.2
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr12_-_323689 | 0.12 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr8_-_25902876 | 0.12 |
ENST00000520164.1
|
EBF2
|
early B-cell factor 2 |
| chr11_+_64323156 | 0.12 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr17_-_33446735 | 0.12 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr14_+_73563735 | 0.12 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr14_+_58711539 | 0.12 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr4_-_156298028 | 0.12 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr14_+_22771851 | 0.12 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chrX_+_56259316 | 0.12 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr11_+_64323098 | 0.12 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr10_-_96122682 | 0.12 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr7_-_141541221 | 0.11 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr1_+_13421176 | 0.11 |
ENST00000376152.1
|
PRAMEF9
|
PRAME family member 9 |
| chr12_-_6798616 | 0.11 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr17_+_32646055 | 0.11 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chrX_-_152864632 | 0.11 |
ENST00000406277.2
|
FAM58A
|
family with sequence similarity 58, member A |
| chr12_-_11036844 | 0.11 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr19_+_46800289 | 0.11 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr17_+_4675175 | 0.11 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chrX_+_8432871 | 0.11 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr9_+_132427883 | 0.11 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr6_+_31554962 | 0.11 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr1_+_87595542 | 0.11 |
ENST00000461990.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr15_+_75550940 | 0.11 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family, member C |
| chr21_-_33957805 | 0.10 |
ENST00000300258.3
ENST00000472557.1 |
TCP10L
|
t-complex 10-like |
| chr9_+_84603687 | 0.10 |
ENST00000344803.2
|
SPATA31D1
|
SPATA31 subfamily D, member 1 |
| chr3_+_44754126 | 0.10 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr2_+_219536749 | 0.10 |
ENST00000295709.3
ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36
|
serine/threonine kinase 36 |
| chr3_+_101818088 | 0.10 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr8_+_94767072 | 0.10 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr16_-_15463926 | 0.10 |
ENST00000432570.2
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chrX_+_144908928 | 0.10 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr1_+_120049826 | 0.10 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr11_-_118436707 | 0.10 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr18_-_11148587 | 0.10 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr6_+_31583761 | 0.10 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr7_+_142498725 | 0.10 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr11_+_125365110 | 0.10 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr13_-_60737898 | 0.10 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr19_+_56116771 | 0.10 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr9_-_79307096 | 0.10 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr7_+_29519486 | 0.09 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr5_-_35195338 | 0.09 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chr6_+_131894284 | 0.09 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr9_+_77230499 | 0.09 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr12_+_56862301 | 0.09 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr14_+_78266408 | 0.09 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr14_+_78266436 | 0.09 |
ENST00000557501.1
ENST00000341211.5 |
ADCK1
|
aarF domain containing kinase 1 |
| chr12_-_5352315 | 0.09 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr12_+_100750846 | 0.09 |
ENST00000323346.5
|
SLC17A8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr1_+_38022572 | 0.09 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr3_-_74570291 | 0.09 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr6_-_150217195 | 0.09 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr4_+_78829479 | 0.09 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr19_-_48389651 | 0.09 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr17_+_35732955 | 0.09 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr16_+_88636789 | 0.08 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr19_+_16222439 | 0.08 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr16_+_84801852 | 0.08 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chrX_+_114524275 | 0.08 |
ENST00000371921.1
ENST00000451986.2 ENST00000371920.3 |
LUZP4
|
leucine zipper protein 4 |
| chrX_+_2670066 | 0.08 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr1_-_12891264 | 0.08 |
ENST00000535591.1
ENST00000437584.1 |
PRAMEF11
|
PRAME family member 11 |
| chr12_-_57882498 | 0.08 |
ENST00000550288.1
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr14_-_24615805 | 0.08 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_-_8113542 | 0.08 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr1_-_67266939 | 0.08 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr9_-_74675521 | 0.08 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr11_+_85339623 | 0.08 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr6_-_52710893 | 0.08 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr12_-_55378452 | 0.08 |
ENST00000449076.1
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr12_-_53594227 | 0.08 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr9_+_140032842 | 0.08 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr10_-_47222824 | 0.08 |
ENST00000355232.3
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr17_-_6915646 | 0.08 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr9_-_6645628 | 0.08 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr12_-_15104040 | 0.07 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr20_+_34824355 | 0.07 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr4_-_156298087 | 0.07 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr10_-_128210005 | 0.07 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr16_-_2581409 | 0.07 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.8 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 1.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.3 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.5 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.7 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.2 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 0.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |