Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
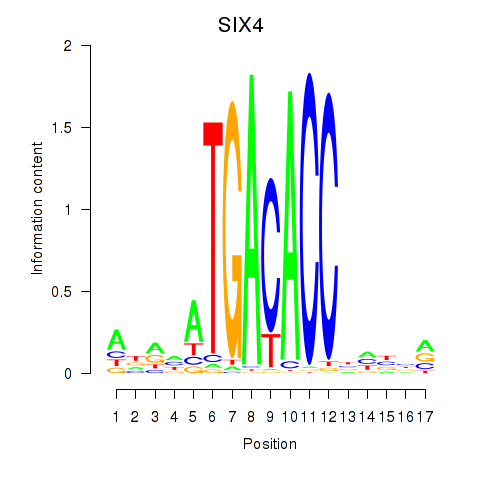
Results for SIX4
Z-value: 0.62
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.8 | SIX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX4 | hg19_v2_chr14_-_61190754_61190852 | 0.59 | 5.3e-04 | Click! |
Activity profile of SIX4 motif
Sorted Z-values of SIX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_207119738 | 4.11 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr1_+_38022572 | 2.43 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr1_-_151778630 | 1.65 |
ENST00000368820.3
|
LINGO4
|
leucine rich repeat and Ig domain containing 4 |
| chr12_-_10282836 | 1.64 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_-_52488048 | 1.49 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chrY_+_22737678 | 1.44 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chrY_+_22737604 | 1.36 |
ENST00000361365.2
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr16_+_640201 | 1.21 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr12_+_133758115 | 1.10 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr12_-_10282742 | 1.05 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_-_10282681 | 1.02 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_+_133757995 | 0.97 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr8_+_144798429 | 0.92 |
ENST00000338033.4
ENST00000395107.4 ENST00000395108.2 |
MAPK15
|
mitogen-activated protein kinase 15 |
| chr3_+_118866222 | 0.91 |
ENST00000490594.1
|
RP11-484M3.5
|
Uncharacterized protein |
| chr19_+_13134772 | 0.81 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_-_130184555 | 0.74 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr14_-_67981916 | 0.71 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr8_+_144295067 | 0.70 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr6_-_76072719 | 0.69 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_+_219135115 | 0.66 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr11_-_130184470 | 0.63 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr11_-_96076334 | 0.61 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr3_+_32147997 | 0.58 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr1_+_35734562 | 0.55 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr1_+_226250379 | 0.54 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr1_-_247335269 | 0.50 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr7_+_69064300 | 0.48 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr6_+_52051171 | 0.48 |
ENST00000340057.1
|
IL17A
|
interleukin 17A |
| chr20_-_52687059 | 0.48 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr22_-_39268192 | 0.44 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr14_-_92413353 | 0.44 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr8_+_95907993 | 0.44 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr6_+_1080164 | 0.43 |
ENST00000314040.1
|
AL033381.1
|
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr2_+_128458514 | 0.42 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr19_+_13135386 | 0.40 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_-_31704282 | 0.39 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr1_+_161123536 | 0.39 |
ENST00000368003.5
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chrX_+_135388147 | 0.38 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr17_-_72527605 | 0.34 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr19_-_474880 | 0.33 |
ENST00000382696.3
ENST00000315489.4 |
ODF3L2
|
outer dense fiber of sperm tails 3-like 2 |
| chr22_-_30960876 | 0.33 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr3_+_32726620 | 0.32 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr1_+_116915855 | 0.32 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_+_231473743 | 0.31 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr2_-_219134822 | 0.31 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr12_+_4829507 | 0.31 |
ENST00000252318.2
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8) |
| chr4_-_13546632 | 0.30 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr14_-_67981870 | 0.30 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr8_-_123139423 | 0.29 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr17_-_61523622 | 0.28 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr2_-_219134343 | 0.28 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr8_+_132916318 | 0.27 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_160097462 | 0.27 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr19_+_19174795 | 0.27 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr9_+_132427883 | 0.26 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr22_-_39268308 | 0.26 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr15_-_20193370 | 0.25 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr1_+_8378140 | 0.23 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr10_+_88414338 | 0.23 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr19_+_35417939 | 0.23 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr4_-_84255935 | 0.23 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr10_+_88414298 | 0.23 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr5_-_1295104 | 0.22 |
ENST00000334602.6
ENST00000508104.2 ENST00000310581.5 ENST00000296820.5 |
TERT
|
telomerase reverse transcriptase |
| chr7_-_121944491 | 0.22 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr19_+_35417844 | 0.22 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr13_+_33160553 | 0.22 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr13_-_78492955 | 0.22 |
ENST00000446573.1
|
EDNRB
|
endothelin receptor type B |
| chr12_-_4488872 | 0.20 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr11_-_18956556 | 0.19 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr1_-_154934200 | 0.19 |
ENST00000368457.2
|
PYGO2
|
pygopus family PHD finger 2 |
| chr11_-_35547572 | 0.19 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr19_-_47128294 | 0.17 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr17_+_7465216 | 0.16 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr12_-_58135903 | 0.16 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_+_156611704 | 0.15 |
ENST00000329117.5
|
BCAN
|
brevican |
| chrX_+_108780347 | 0.14 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr7_+_69064566 | 0.13 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr15_+_41245160 | 0.13 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr4_-_2420335 | 0.12 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chrX_+_54466829 | 0.12 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chrX_-_119763835 | 0.11 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr1_+_156611960 | 0.10 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr10_+_26505179 | 0.09 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr1_+_26737253 | 0.08 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr1_-_114414316 | 0.08 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr3_-_183967296 | 0.08 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr8_+_56014949 | 0.06 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr17_+_56232494 | 0.06 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr1_+_26737292 | 0.05 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr10_+_26505594 | 0.05 |
ENST00000259271.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr6_-_41040195 | 0.05 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr10_+_7860460 | 0.05 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr11_+_2415061 | 0.04 |
ENST00000481687.1
|
CD81
|
CD81 molecule |
| chr7_+_97361218 | 0.04 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr5_-_78365437 | 0.04 |
ENST00000380311.4
ENST00000540686.1 ENST00000255189.3 |
DMGDH
|
dimethylglycine dehydrogenase |
| chr20_+_57875457 | 0.04 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr7_-_99063769 | 0.04 |
ENST00000394186.3
ENST00000359832.4 ENST00000449683.1 ENST00000488775.1 ENST00000523680.1 ENST00000292475.3 ENST00000430982.1 ENST00000555673.1 ENST00000413834.1 |
ATP5J2
PTCD1
ATP5J2-PTCD1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 pentatricopeptide repeat domain 1 ATP5J2-PTCD1 readthrough |
| chrX_+_56100757 | 0.03 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chr11_+_7273181 | 0.03 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chr3_-_49158218 | 0.03 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr7_+_97361388 | 0.03 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr3_+_72200408 | 0.03 |
ENST00000473713.1
|
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr2_-_180427304 | 0.02 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr1_-_231473578 | 0.02 |
ENST00000360394.2
ENST00000366645.1 |
EXOC8
|
exocyst complex component 8 |
| chr3_-_87325612 | 0.02 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr20_+_57875658 | 0.01 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr1_-_42800860 | 0.01 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chr14_-_106692191 | 0.01 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr5_+_85913721 | 0.01 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr2_+_120687335 | 0.01 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr17_-_39684550 | 0.01 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr15_+_59279851 | 0.01 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr11_-_5364809 | 0.01 |
ENST00000300773.2
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5 |
| chr7_-_26578407 | 0.00 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 2.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.4 | 1.5 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 3.7 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.3 | GO:0015880 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.2 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.0 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 4.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 2.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.2 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 3.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.7 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.2 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.2 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.6 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |