Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
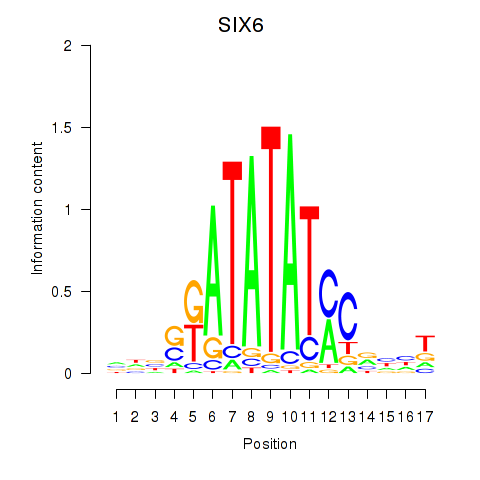
Results for SIX6
Z-value: 0.54
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX6 | hg19_v2_chr14_+_60975644_60975673 | 0.30 | 1.1e-01 | Click! |
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_80263506 | 1.22 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr12_-_28122980 | 0.91 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr2_-_88427568 | 0.87 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr12_-_121477039 | 0.84 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr8_+_67405438 | 0.80 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr12_-_28123206 | 0.71 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr12_-_121476959 | 0.70 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr3_-_111314230 | 0.67 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr3_+_8543393 | 0.65 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr6_+_146348782 | 0.64 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr19_-_44031375 | 0.56 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr9_-_21142144 | 0.54 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr6_+_146348810 | 0.52 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr12_+_119616447 | 0.51 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr3_+_8543561 | 0.49 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr12_-_14849470 | 0.48 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr2_-_239140011 | 0.45 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr22_-_19137796 | 0.45 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr8_+_1993152 | 0.41 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr6_-_26285737 | 0.40 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr13_-_20080080 | 0.39 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr8_+_1993173 | 0.39 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr10_+_91061712 | 0.39 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr8_+_67104323 | 0.37 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr14_+_20937538 | 0.37 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr12_-_71512027 | 0.33 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein |
| chr22_+_18721427 | 0.33 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr12_-_10978957 | 0.32 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr19_-_14628645 | 0.32 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_+_63063152 | 0.32 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr1_-_36235559 | 0.31 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr1_+_86934526 | 0.31 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr21_-_19775973 | 0.30 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr1_+_21880560 | 0.30 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr14_+_21510385 | 0.29 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr9_-_100707116 | 0.29 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chr1_-_161193349 | 0.29 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr22_-_17302589 | 0.28 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr15_+_58430567 | 0.27 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr16_-_87350970 | 0.26 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr6_-_108145499 | 0.26 |
ENST00000369020.3
ENST00000369022.2 |
SCML4
|
sex comb on midleg-like 4 (Drosophila) |
| chr19_+_46000479 | 0.25 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr19_-_1650666 | 0.24 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr5_+_40909354 | 0.24 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr16_+_72090053 | 0.24 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr13_+_34392200 | 0.24 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr15_+_58430368 | 0.24 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr7_+_117251671 | 0.23 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr5_-_78281775 | 0.23 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr4_-_71532339 | 0.23 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_-_124553437 | 0.23 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr6_-_26043885 | 0.23 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr9_-_23821842 | 0.22 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr4_+_88532028 | 0.22 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr14_-_46185155 | 0.21 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr13_-_46679185 | 0.21 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr18_-_69246167 | 0.21 |
ENST00000566582.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr1_-_161015752 | 0.21 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chrX_-_18238999 | 0.20 |
ENST00000380033.4
ENST00000380030.3 |
BEND2
|
BEN domain containing 2 |
| chr4_-_123377880 | 0.20 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr11_+_74699703 | 0.20 |
ENST00000529024.1
ENST00000544263.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr8_+_26247878 | 0.20 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr2_+_85822857 | 0.20 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chrY_-_16098393 | 0.20 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chr13_-_46679144 | 0.20 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr2_+_89184868 | 0.19 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr9_-_21351377 | 0.19 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr12_-_121476750 | 0.18 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr17_-_67057114 | 0.18 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_+_145413268 | 0.18 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr18_-_69246186 | 0.18 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr2_-_134326009 | 0.17 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr3_+_151531810 | 0.17 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chrY_+_16168097 | 0.17 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr10_-_61720640 | 0.17 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr4_-_13546632 | 0.16 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr5_-_177207634 | 0.16 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr6_+_47749718 | 0.15 |
ENST00000489301.2
ENST00000371211.2 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr2_-_228497888 | 0.15 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr9_-_67786624 | 0.14 |
ENST00000455764.2
|
FAM27E3
|
family with sequence similarity 27, member E3 |
| chr3_-_196242233 | 0.14 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr22_+_18043133 | 0.14 |
ENST00000327451.6
ENST00000399813.1 |
SLC25A18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr2_-_208989225 | 0.14 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chrX_+_8433376 | 0.13 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr7_-_141957847 | 0.13 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr1_-_159684371 | 0.13 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr9_-_100684845 | 0.12 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr5_+_55147205 | 0.12 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr1_+_112016414 | 0.12 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr17_-_67057047 | 0.12 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr2_+_62132781 | 0.12 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr1_+_160709076 | 0.12 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr7_-_44530479 | 0.11 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr11_+_30253410 | 0.11 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chrX_-_77225135 | 0.11 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr19_-_16008880 | 0.10 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr10_+_81891416 | 0.10 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr6_+_26204825 | 0.10 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr2_+_62132800 | 0.09 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr16_-_52119019 | 0.09 |
ENST00000561513.1
ENST00000565742.1 |
LINC00919
|
long intergenic non-protein coding RNA 919 |
| chr4_-_70826725 | 0.09 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr14_+_21525981 | 0.09 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chrY_-_24038660 | 0.09 |
ENST00000382677.3
|
RBMY1D
|
RNA binding motif protein, Y-linked, family 1, member D |
| chr4_+_113739244 | 0.09 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr18_+_55018044 | 0.08 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr16_-_20364122 | 0.08 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr8_+_84824920 | 0.08 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr9_-_139642968 | 0.08 |
ENST00000341206.4
|
LCN6
|
lipocalin 6 |
| chr10_-_75008620 | 0.08 |
ENST00000372950.4
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr6_-_26018007 | 0.08 |
ENST00000244573.3
|
HIST1H1A
|
histone cluster 1, H1a |
| chr11_+_122709200 | 0.08 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr1_+_40942887 | 0.08 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr1_+_159557607 | 0.07 |
ENST00000255040.2
|
APCS
|
amyloid P component, serum |
| chr4_-_139051839 | 0.07 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr1_+_160709055 | 0.07 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr15_+_69857515 | 0.07 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chrX_-_53350522 | 0.07 |
ENST00000396435.3
ENST00000375368.5 |
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chrY_+_23698778 | 0.06 |
ENST00000303902.5
|
RBMY1A1
|
RNA binding motif protein, Y-linked, family 1, member A1 |
| chrX_-_118699325 | 0.06 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr1_-_182369751 | 0.06 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr2_-_77749474 | 0.06 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr19_+_3136115 | 0.06 |
ENST00000262958.3
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr11_+_59807748 | 0.06 |
ENST00000278855.2
ENST00000532905.1 |
PLAC1L
|
oocyte secreted protein 2 |
| chr10_+_45495898 | 0.05 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr1_-_150669604 | 0.05 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr6_+_37012607 | 0.05 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr1_-_13673511 | 0.05 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr1_+_234509186 | 0.05 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr12_+_40787194 | 0.05 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr5_+_175490540 | 0.05 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr18_+_50278430 | 0.05 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr16_-_20364030 | 0.04 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr1_+_31885963 | 0.04 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr1_-_157014865 | 0.04 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_-_107596910 | 0.03 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr9_-_14910990 | 0.03 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr20_+_54987168 | 0.03 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr5_+_115420688 | 0.02 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr12_-_122018859 | 0.02 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr20_+_54987305 | 0.02 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr6_-_26189304 | 0.01 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr3_-_53916202 | 0.01 |
ENST00000335754.3
|
ACTR8
|
ARP8 actin-related protein 8 homolog (yeast) |
| chr14_+_22948510 | 0.00 |
ENST00000390483.1
|
TRAJ56
|
T cell receptor alpha joining 56 |
| chr16_-_21868978 | 0.00 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr19_+_55235969 | 0.00 |
ENST00000402254.2
ENST00000538269.1 ENST00000541392.1 ENST00000291860.1 ENST00000396284.2 |
KIR3DL1
KIR3DL3
KIR2DL4
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.3 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.6 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.2 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 1.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |