Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
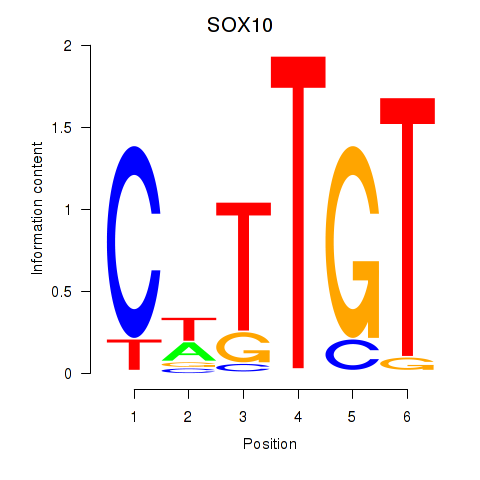
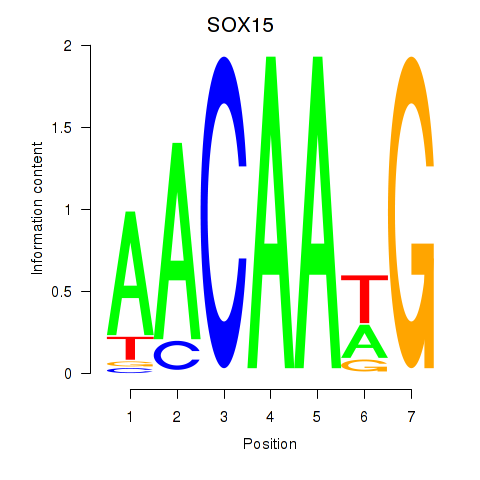
Results for SOX10_SOX15
Z-value: 0.37
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | 0.14 | 4.8e-01 | Click! |
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.01 | 9.7e-01 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_86564776 | 0.62 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr2_-_118943930 | 0.54 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr3_-_123339418 | 0.42 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr15_+_84115868 | 0.42 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr3_-_123339343 | 0.42 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr6_-_127840336 | 0.39 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr2_+_10262857 | 0.39 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr4_+_154387480 | 0.36 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr11_+_125496619 | 0.33 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_200992827 | 0.31 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr6_-_127840021 | 0.29 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chrY_+_15016013 | 0.27 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr7_-_99869799 | 0.27 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr3_+_39509163 | 0.27 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr3_-_141747950 | 0.26 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_157002775 | 0.25 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr6_-_139613269 | 0.25 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr3_+_111260856 | 0.25 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr19_-_51472823 | 0.25 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr7_-_121944491 | 0.24 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr1_+_47799542 | 0.23 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr6_-_28411241 | 0.22 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chrX_-_45629661 | 0.21 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr17_+_75447326 | 0.21 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr14_-_57272366 | 0.20 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr12_-_42631529 | 0.20 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr22_-_23484246 | 0.20 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr3_+_39509070 | 0.19 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr18_-_74728998 | 0.19 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr11_-_94965667 | 0.19 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr2_-_166060552 | 0.19 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr2_+_162272605 | 0.18 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr3_+_141105235 | 0.18 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr12_+_104359641 | 0.18 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr2_-_166060571 | 0.18 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr6_-_42419649 | 0.18 |
ENST00000372922.4
ENST00000541110.1 ENST00000372917.4 |
TRERF1
|
transcriptional regulating factor 1 |
| chr2_+_27237615 | 0.17 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr17_+_7590734 | 0.17 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr5_+_137514834 | 0.16 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr5_+_137514687 | 0.16 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr11_+_86106208 | 0.16 |
ENST00000528728.1
|
CCDC81
|
coiled-coil domain containing 81 |
| chr14_+_97263641 | 0.16 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr2_-_47572105 | 0.16 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr15_+_63335899 | 0.15 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr1_-_53793584 | 0.15 |
ENST00000354412.3
ENST00000347547.2 ENST00000306052.6 |
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_-_9940058 | 0.15 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chrX_+_150869023 | 0.15 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr2_-_10588630 | 0.15 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr2_-_1748214 | 0.15 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr8_+_81398444 | 0.14 |
ENST00000455036.3
ENST00000426744.2 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr12_-_8815299 | 0.14 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_49582593 | 0.14 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_151986709 | 0.14 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr12_-_8815215 | 0.14 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr6_+_30689350 | 0.14 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr4_-_123377880 | 0.14 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr6_+_72596604 | 0.14 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr6_-_27880174 | 0.14 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr7_-_111846435 | 0.13 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_-_66445219 | 0.13 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr17_-_56406117 | 0.13 |
ENST00000268893.6
ENST00000355701.3 |
BZRAP1
|
benzodiazepine receptor (peripheral) associated protein 1 |
| chrX_+_129473859 | 0.13 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr22_+_39052632 | 0.13 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr15_+_84116106 | 0.13 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr1_-_79472365 | 0.13 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr15_-_88799948 | 0.13 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr11_+_128563652 | 0.13 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_157002749 | 0.12 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr13_+_30002741 | 0.12 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr11_-_10829851 | 0.12 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chrX_-_117107542 | 0.12 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr5_-_138718973 | 0.12 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr6_-_112575838 | 0.12 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr2_-_188312971 | 0.12 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr3_-_167813132 | 0.12 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_+_174417095 | 0.12 |
ENST00000367685.2
|
GPR52
|
G protein-coupled receptor 52 |
| chr17_-_7297519 | 0.11 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_+_215256467 | 0.11 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr7_-_148581251 | 0.11 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr5_-_65017921 | 0.11 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr19_+_47105309 | 0.11 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr13_-_24007815 | 0.11 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr18_+_6729698 | 0.11 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr7_+_107110488 | 0.11 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr1_-_26233423 | 0.11 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr1_+_240255166 | 0.11 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr10_-_97321165 | 0.11 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr5_+_60933634 | 0.11 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr3_-_71353892 | 0.10 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr6_-_161695042 | 0.10 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr16_+_89989687 | 0.10 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr4_-_138453606 | 0.10 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr9_-_120177342 | 0.10 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr14_-_51027838 | 0.10 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_161695074 | 0.10 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr17_-_7297833 | 0.10 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chrX_+_36254051 | 0.10 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr6_-_134861089 | 0.10 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr11_+_111808119 | 0.10 |
ENST00000531396.1
|
DIXDC1
|
DIX domain containing 1 |
| chr11_-_5364809 | 0.10 |
ENST00000300773.2
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5 |
| chr12_-_11508520 | 0.09 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr8_+_67039131 | 0.09 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr11_+_111807863 | 0.09 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr8_-_9760839 | 0.09 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr7_+_154002189 | 0.09 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr20_+_44036620 | 0.09 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_-_30654977 | 0.09 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr8_+_67039278 | 0.09 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr20_-_4804244 | 0.09 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr11_-_89224488 | 0.09 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr17_-_49124230 | 0.09 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr7_-_94953878 | 0.09 |
ENST00000222381.3
|
PON1
|
paraoxonase 1 |
| chr1_-_6479963 | 0.09 |
ENST00000377836.4
ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2
|
hes family bHLH transcription factor 2 |
| chr11_-_89224299 | 0.09 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr17_+_36584662 | 0.09 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr12_-_11548496 | 0.09 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr17_-_18950310 | 0.09 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr3_-_167813672 | 0.09 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr16_+_84801852 | 0.09 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr2_-_26205340 | 0.09 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr7_+_134551583 | 0.08 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr16_+_27078219 | 0.08 |
ENST00000418886.1
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr1_+_24117627 | 0.08 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr1_+_197886461 | 0.08 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr10_+_24528108 | 0.08 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr12_-_11463353 | 0.08 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr15_-_50647370 | 0.08 |
ENST00000558970.1
ENST00000396464.3 ENST00000560825.1 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr19_+_34287174 | 0.08 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr17_-_79849438 | 0.08 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr17_+_67498538 | 0.08 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr14_-_71107921 | 0.08 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr2_-_153573965 | 0.08 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr15_-_70388943 | 0.08 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr2_+_153191706 | 0.08 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr11_+_67007518 | 0.08 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr8_-_87755878 | 0.07 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr6_+_30689401 | 0.07 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr11_-_89224508 | 0.07 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr1_+_11751748 | 0.07 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr20_+_20348740 | 0.07 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr7_-_150924121 | 0.07 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr12_+_15475462 | 0.07 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr6_-_165989936 | 0.07 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr15_+_92937058 | 0.07 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr14_-_50583271 | 0.07 |
ENST00000395860.2
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine (K) methyltransferase |
| chr17_+_75283973 | 0.07 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr3_+_111260954 | 0.07 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr5_-_150521192 | 0.07 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr15_-_50647347 | 0.07 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr4_+_95128748 | 0.07 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_9005917 | 0.07 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr12_-_95611149 | 0.07 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr22_-_21984282 | 0.07 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr12_+_26111823 | 0.07 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr15_-_88799384 | 0.07 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chrX_-_46187069 | 0.07 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_+_31477250 | 0.07 |
ENST00000313737.4
|
AC024940.1
|
AC024940.1 |
| chr8_+_81397846 | 0.07 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr4_+_95129061 | 0.07 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr20_+_60698180 | 0.06 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chrX_-_13956497 | 0.06 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr17_-_48072574 | 0.06 |
ENST00000434704.2
|
DLX3
|
distal-less homeobox 3 |
| chr13_+_111767650 | 0.06 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_-_24665353 | 0.06 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr19_+_10765699 | 0.06 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr4_-_5890145 | 0.06 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr7_+_23145884 | 0.06 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr15_-_50647274 | 0.06 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr3_-_4927447 | 0.06 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr15_+_58724184 | 0.06 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr6_-_91006461 | 0.06 |
ENST00000257749.4
ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr20_-_22559211 | 0.06 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr2_-_161350305 | 0.06 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_-_209028300 | 0.06 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr7_-_27219849 | 0.05 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr17_+_64961026 | 0.05 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr15_-_88799661 | 0.05 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr6_-_112575912 | 0.05 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr12_-_90049878 | 0.05 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_-_80994210 | 0.05 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr12_-_90049828 | 0.05 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_+_95128996 | 0.05 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_119031228 | 0.05 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr13_+_98795505 | 0.05 |
ENST00000319562.6
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr13_+_78315466 | 0.05 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_98795434 | 0.05 |
ENST00000376586.2
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr7_-_27142290 | 0.05 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr15_-_56035177 | 0.05 |
ENST00000389286.4
ENST00000561292.1 |
PRTG
|
protogenin |
| chr18_+_32558208 | 0.05 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_69064566 | 0.05 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr17_-_46690839 | 0.05 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr6_+_114178512 | 0.05 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr21_+_30671690 | 0.05 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chrX_-_142722897 | 0.05 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr5_+_38148582 | 0.05 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr12_+_113495492 | 0.05 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr20_-_30795511 | 0.05 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr8_+_81397876 | 0.05 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr2_+_32288657 | 0.05 |
ENST00000345662.1
|
SPAST
|
spastin |
| chrX_-_63425561 | 0.05 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0072376 | blood coagulation, intrinsic pathway(GO:0007597) protein activation cascade(GO:0072376) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |