Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
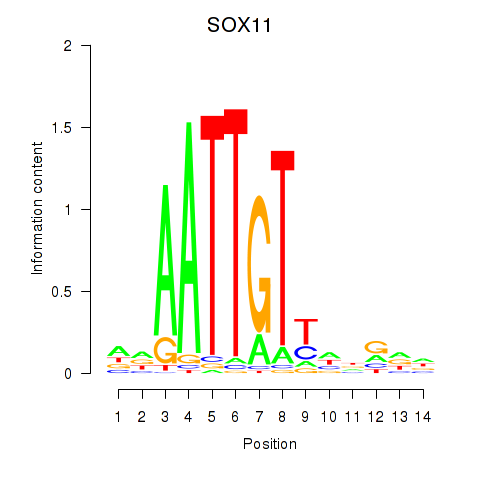
Results for SOX11
Z-value: 0.65
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | -0.20 | 3.0e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_7921090 | 6.34 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr6_-_32557610 | 2.98 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr11_+_61976137 | 2.83 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr14_+_96949319 | 2.59 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr3_-_112564797 | 2.03 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr8_-_128960591 | 1.48 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr12_+_56075330 | 1.36 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr5_+_140213815 | 1.27 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr10_-_25305011 | 1.24 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr4_+_159443090 | 1.20 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr5_+_140248518 | 1.20 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr1_+_244515930 | 1.16 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr4_+_72204755 | 1.11 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr6_+_159071015 | 1.01 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr6_-_76072719 | 1.00 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_-_105416039 | 0.98 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chrX_-_132231123 | 0.98 |
ENST00000511190.1
|
USP26
|
ubiquitin specific peptidase 26 |
| chr2_+_42104692 | 0.91 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr6_-_52705641 | 0.86 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr6_+_131958436 | 0.76 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr5_-_160279207 | 0.75 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr3_+_113616317 | 0.74 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr2_-_225266743 | 0.72 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr20_+_18794370 | 0.68 |
ENST00000377428.2
|
SCP2D1
|
SCP2 sterol-binding domain containing 1 |
| chr2_+_169658928 | 0.67 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr8_+_104831554 | 0.66 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_169659121 | 0.65 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr8_-_72274095 | 0.63 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr2_-_225266711 | 0.61 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr6_-_116381918 | 0.58 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr1_-_120935894 | 0.58 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr8_+_105352050 | 0.57 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr2_+_47596287 | 0.56 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr3_+_98072698 | 0.55 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor, family 5, subfamily K, member 4 |
| chr17_-_34344991 | 0.54 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr7_-_92855762 | 0.51 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_-_34345002 | 0.50 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr10_+_32873190 | 0.50 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr8_+_118147498 | 0.50 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr13_-_30424821 | 0.48 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr15_+_49447947 | 0.47 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr19_-_14911023 | 0.47 |
ENST00000248073.2
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1 |
| chr3_+_69812701 | 0.47 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_-_25268104 | 0.44 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr14_+_21156915 | 0.42 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr2_+_27799389 | 0.42 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr11_+_4664650 | 0.41 |
ENST00000396952.5
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr6_-_49712123 | 0.41 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr1_+_196621156 | 0.40 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr10_+_118350522 | 0.39 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr1_-_197169672 | 0.38 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr3_-_49967292 | 0.38 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr7_-_122635754 | 0.37 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr15_-_37392086 | 0.36 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr4_+_130017268 | 0.36 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr6_-_49712147 | 0.35 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr13_+_78315466 | 0.34 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_127505167 | 0.33 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr11_+_92085262 | 0.33 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr7_-_112635675 | 0.33 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr2_+_109237717 | 0.32 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_+_67381263 | 0.32 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr7_-_7575477 | 0.32 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr11_+_22688150 | 0.31 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr2_+_138721850 | 0.29 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr16_+_67381289 | 0.28 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr5_-_54468974 | 0.28 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr2_-_178128250 | 0.28 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr6_-_154751629 | 0.27 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr7_-_56118981 | 0.27 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr1_-_150669500 | 0.27 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_+_24241789 | 0.27 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr17_+_3118915 | 0.25 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr1_+_204839959 | 0.25 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr2_+_169757750 | 0.24 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr9_+_71986182 | 0.24 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr11_+_7110165 | 0.23 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr15_+_45028753 | 0.23 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr7_-_122342966 | 0.23 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr7_-_122342988 | 0.22 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr19_+_50094866 | 0.22 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr3_+_112930306 | 0.20 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr5_+_140207536 | 0.19 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr2_+_207804278 | 0.19 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr12_-_122879969 | 0.19 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr16_+_67261008 | 0.19 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr6_-_136610911 | 0.19 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr2_+_166326157 | 0.18 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr8_+_24241969 | 0.17 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chrX_-_13835147 | 0.17 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr14_+_39703112 | 0.17 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr3_+_113251143 | 0.17 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr6_-_46048116 | 0.17 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr4_+_110749143 | 0.17 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr10_-_5446786 | 0.17 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr14_-_36789865 | 0.17 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr14_-_36789783 | 0.17 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr6_-_11779014 | 0.16 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_31125850 | 0.16 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr1_+_145524891 | 0.16 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chrX_+_130192318 | 0.16 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr6_-_99873145 | 0.16 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr15_+_45028719 | 0.16 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr4_-_170679024 | 0.16 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr1_+_65730385 | 0.15 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_+_95972822 | 0.15 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_196621002 | 0.15 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr13_+_78315295 | 0.15 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr11_-_58612168 | 0.15 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr6_-_11779174 | 0.15 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr2_-_71222466 | 0.15 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr20_-_30539773 | 0.15 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr9_+_27109133 | 0.14 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chrX_+_27826107 | 0.14 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr12_-_81763127 | 0.13 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_-_58611957 | 0.13 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr8_+_19796381 | 0.13 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr8_-_25281747 | 0.13 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr7_-_82792215 | 0.13 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr20_-_32308028 | 0.13 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr3_-_167191814 | 0.12 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr2_-_20251744 | 0.12 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr7_-_81399329 | 0.10 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr19_+_39989535 | 0.10 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr11_-_95657231 | 0.10 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr5_-_86534822 | 0.10 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr1_-_241799232 | 0.10 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr1_+_152943122 | 0.10 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr1_+_149754227 | 0.09 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr3_-_18466026 | 0.08 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr5_-_169725231 | 0.08 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr17_-_10017864 | 0.07 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr2_+_102927962 | 0.07 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr6_-_133055815 | 0.07 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr11_+_57425209 | 0.06 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr5_+_172571445 | 0.06 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr10_+_35456444 | 0.06 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr17_-_39324424 | 0.05 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr12_+_50794592 | 0.05 |
ENST00000293618.8
ENST00000429001.3 ENST00000548174.1 ENST00000548697.1 ENST00000548993.1 ENST00000398473.2 ENST00000522085.1 ENST00000518444.1 ENST00000551886.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr18_+_7754957 | 0.05 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_+_141294628 | 0.04 |
ENST00000512749.1
ENST00000608372.1 ENST00000506597.1 ENST00000394201.4 ENST00000510586.1 |
SCOC
|
short coiled-coil protein |
| chr15_+_69857515 | 0.04 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr8_+_9413410 | 0.04 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr4_-_73434498 | 0.04 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr4_+_183164574 | 0.04 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chrM_+_7586 | 0.04 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr6_-_47277634 | 0.03 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr13_-_103019744 | 0.03 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr15_-_99791413 | 0.03 |
ENST00000394129.2
ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr3_-_133380731 | 0.03 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr18_+_46065483 | 0.02 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr6_+_30585486 | 0.02 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr9_-_95166841 | 0.02 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr6_-_36515177 | 0.01 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr1_+_214776516 | 0.01 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr21_-_37852359 | 0.01 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr1_+_198608146 | 0.01 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr2_-_176046391 | 0.01 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr4_+_71337834 | 0.01 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr3_+_178253993 | 0.00 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chrM_+_8527 | 0.00 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chrX_+_102192200 | 0.00 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 3.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.3 | 0.8 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 0.6 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 2.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 2.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.3 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 1.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 6.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 1.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.4 | 2.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 2.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 6.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |