Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
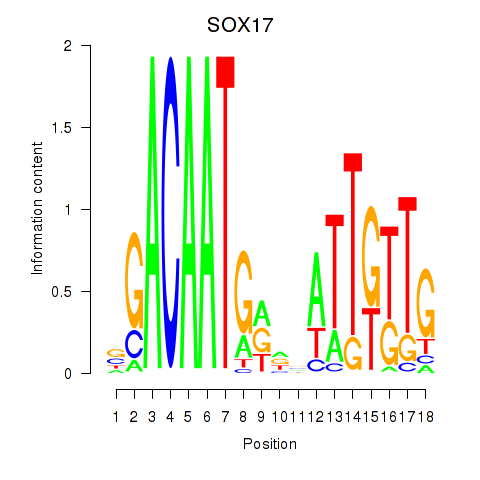
Results for SOX17
Z-value: 0.89
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg19_v2_chr8_+_55370487_55370503 | -0.61 | 3.5e-04 | Click! |
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_71533055 | 9.64 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chrY_+_2709906 | 4.19 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr8_+_72587535 | 4.00 |
ENST00000519840.1
ENST00000521131.1 |
RP11-1144P22.1
|
RP11-1144P22.1 |
| chr13_-_39564993 | 3.73 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr6_-_32920794 | 3.54 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr3_-_19975665 | 3.38 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr17_+_41158742 | 2.55 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr14_-_93673353 | 2.49 |
ENST00000556566.1
ENST00000306954.4 |
C14orf142
|
chromosome 14 open reading frame 142 |
| chrX_-_38186811 | 2.42 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr6_-_52668605 | 2.27 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr3_-_167371740 | 2.10 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr5_-_16509101 | 2.08 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_+_68100989 | 2.08 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_52710893 | 2.04 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr8_+_110098850 | 2.03 |
ENST00000518632.1
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr2_+_233527443 | 1.82 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr19_-_10764509 | 1.71 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr10_-_69597915 | 1.62 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_-_15374343 | 1.61 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr5_+_68860949 | 1.54 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr4_-_100212132 | 1.50 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr5_-_41213607 | 1.47 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr11_-_108464465 | 1.45 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr3_-_52488048 | 1.44 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chrX_-_55187531 | 1.38 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr16_+_71560023 | 1.36 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr4_-_76008706 | 1.36 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr6_+_159071015 | 1.33 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr13_+_24144796 | 1.33 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_+_22694606 | 1.31 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr3_+_125687987 | 1.30 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr11_+_17316870 | 1.28 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr9_+_86237963 | 1.27 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr1_+_15256230 | 1.26 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr1_+_171217622 | 1.24 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chrX_-_55187588 | 1.24 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr2_-_38303218 | 1.23 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr8_+_110099653 | 1.21 |
ENST00000311762.2
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr1_+_171283331 | 1.20 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr6_-_52628271 | 1.20 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr5_-_159827073 | 1.18 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr2_-_27851843 | 1.17 |
ENST00000324364.3
|
CCDC121
|
coiled-coil domain containing 121 |
| chr2_-_157198860 | 1.17 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_-_9006766 | 1.17 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr1_-_48937838 | 1.16 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr12_+_55248289 | 1.15 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr18_-_19748331 | 1.14 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr19_-_9003586 | 1.13 |
ENST00000380951.5
|
MUC16
|
mucin 16, cell surface associated |
| chr19_+_58514229 | 1.13 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr15_+_88120158 | 1.11 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr5_-_159827033 | 1.09 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr1_+_227751231 | 1.09 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr1_+_174844645 | 1.08 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_111358372 | 1.08 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr2_-_159237472 | 1.07 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr5_-_38595498 | 1.04 |
ENST00000263409.4
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr6_-_52705641 | 1.02 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr1_+_226013047 | 1.02 |
ENST00000366837.4
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr1_+_43312258 | 1.01 |
ENST00000372508.3
ENST00000372507.1 ENST00000372506.1 ENST00000397044.3 ENST00000372504.1 |
ZNF691
|
zinc finger protein 691 |
| chr4_-_168155730 | 0.99 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_56621665 | 0.95 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr1_-_48937821 | 0.91 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr12_-_40499661 | 0.90 |
ENST00000280871.4
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13 |
| chr6_+_26217159 | 0.88 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr20_-_45318230 | 0.87 |
ENST00000372114.3
|
TP53RK
|
TP53 regulating kinase |
| chr16_-_71523179 | 0.83 |
ENST00000564230.1
ENST00000565637.1 |
ZNF19
|
zinc finger protein 19 |
| chr6_-_31745085 | 0.82 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr6_-_31239846 | 0.82 |
ENST00000415537.1
ENST00000376228.5 ENST00000383329.3 |
HLA-C
|
major histocompatibility complex, class I, C |
| chr12_+_69742121 | 0.82 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr6_-_31745037 | 0.82 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr11_-_3818688 | 0.81 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr14_-_70883708 | 0.81 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr10_+_53806501 | 0.81 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr16_+_29467780 | 0.81 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr11_-_110561721 | 0.80 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr12_+_104680659 | 0.76 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr3_-_49170522 | 0.76 |
ENST00000418109.1
|
LAMB2
|
laminin, beta 2 (laminin S) |
| chr2_-_118943930 | 0.75 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr17_+_76494911 | 0.75 |
ENST00000598378.1
|
DNAH17-AS1
|
DNAH17 antisense RNA 1 |
| chr16_-_30023615 | 0.74 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr6_-_119256311 | 0.74 |
ENST00000316316.6
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr1_+_174417095 | 0.73 |
ENST00000367685.2
|
GPR52
|
G protein-coupled receptor 52 |
| chr3_-_3152031 | 0.72 |
ENST00000383846.1
ENST00000427088.1 ENST00000446632.2 ENST00000438560.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr19_-_19843900 | 0.71 |
ENST00000344099.3
|
ZNF14
|
zinc finger protein 14 |
| chr3_-_49170405 | 0.70 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr16_-_71523236 | 0.70 |
ENST00000288177.5
ENST00000569072.1 |
ZNF19
|
zinc finger protein 19 |
| chr15_-_81202118 | 0.69 |
ENST00000560560.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr19_-_19887185 | 0.68 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr17_-_56605341 | 0.67 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr15_+_34310428 | 0.65 |
ENST00000557872.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr4_-_87028478 | 0.65 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chrX_+_129473916 | 0.64 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr12_-_57443886 | 0.64 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr16_+_16484691 | 0.64 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr19_+_6135646 | 0.64 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr2_-_100939195 | 0.62 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr6_+_32605134 | 0.62 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr10_-_27389392 | 0.61 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr17_-_58042075 | 0.61 |
ENST00000305783.8
ENST00000589113.1 ENST00000442346.2 |
RNFT1
|
ring finger protein, transmembrane 1 |
| chr19_+_49956426 | 0.60 |
ENST00000293350.4
ENST00000540132.1 ENST00000455361.2 ENST00000433981.2 |
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr7_+_139026057 | 0.60 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr1_+_59775752 | 0.59 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr16_-_15474904 | 0.59 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr8_-_95220775 | 0.59 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr6_+_29910301 | 0.58 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr3_+_46412345 | 0.58 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chrX_+_49216659 | 0.58 |
ENST00000415752.1
|
GAGE12I
|
G antigen 12I |
| chr3_-_123710893 | 0.58 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr6_+_28109703 | 0.56 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr15_-_20193370 | 0.56 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr4_-_168155577 | 0.56 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_-_24895566 | 0.55 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr2_+_232573222 | 0.55 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr3_-_151176497 | 0.55 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr3_-_11610255 | 0.55 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr5_+_49962772 | 0.55 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_+_25205568 | 0.54 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr2_+_86668464 | 0.54 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr17_-_67264947 | 0.54 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr21_+_29911640 | 0.53 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr6_-_26659913 | 0.53 |
ENST00000480036.1
ENST00000415922.2 |
ZNF322
|
zinc finger protein 322 |
| chr4_-_150736962 | 0.52 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr20_-_43883197 | 0.52 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr1_+_40840320 | 0.52 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr20_-_44168046 | 0.52 |
ENST00000372665.3
ENST00000372670.3 ENST00000600168.1 |
WFDC6
|
WAP four-disulfide core domain 6 |
| chr19_-_48752812 | 0.52 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr4_-_168155700 | 0.51 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_205225319 | 0.50 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr10_-_115614127 | 0.50 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr19_-_44809121 | 0.49 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr8_+_133931648 | 0.49 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr1_-_160492994 | 0.49 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr11_-_123756334 | 0.48 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr6_-_152958521 | 0.48 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr13_+_103459704 | 0.48 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr9_-_115818951 | 0.47 |
ENST00000553380.1
ENST00000374227.3 |
ZFP37
|
ZFP37 zinc finger protein |
| chr19_-_48753104 | 0.47 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr6_-_42690312 | 0.47 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr11_+_65266507 | 0.47 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr5_+_125758813 | 0.46 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_-_219906220 | 0.46 |
ENST00000458526.1
ENST00000409865.3 ENST00000410037.1 ENST00000457968.1 ENST00000436631.1 ENST00000341552.5 ENST00000441968.1 ENST00000295729.2 |
CCDC108
|
coiled-coil domain containing 108 |
| chr3_+_174577070 | 0.46 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr6_-_119031228 | 0.46 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr10_+_6622345 | 0.46 |
ENST00000445427.1
ENST00000455810.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr9_-_125027079 | 0.45 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr12_+_25205666 | 0.45 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr13_+_111766897 | 0.45 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_+_170581213 | 0.45 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr8_+_21916710 | 0.44 |
ENST00000523266.1
ENST00000519907.1 |
DMTN
|
dematin actin binding protein |
| chr3_-_46000064 | 0.44 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr6_+_1080164 | 0.44 |
ENST00000314040.1
|
AL033381.1
|
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr1_-_45805667 | 0.43 |
ENST00000488731.2
ENST00000435155.1 |
MUTYH
|
mutY homolog |
| chr18_+_5238055 | 0.43 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr1_-_45805607 | 0.43 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr11_+_3819049 | 0.43 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr12_-_55367361 | 0.43 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr12_-_90024360 | 0.43 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_+_498500 | 0.43 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr11_-_35440579 | 0.43 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_+_24857552 | 0.43 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr5_-_10308125 | 0.42 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chrX_-_8139308 | 0.42 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr19_+_34895289 | 0.41 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr5_+_125758865 | 0.41 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_+_22333943 | 0.41 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr16_+_28770025 | 0.40 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr6_+_126277842 | 0.39 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr14_-_68066975 | 0.38 |
ENST00000559581.1
ENST00000560722.1 ENST00000559415.1 ENST00000216452.4 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr19_+_32836499 | 0.38 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr19_+_34745442 | 0.38 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chrX_+_49363665 | 0.38 |
ENST00000381700.6
|
GAGE1
|
G antigen 1 |
| chr10_-_135379132 | 0.37 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr1_+_95975672 | 0.37 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr3_+_88108381 | 0.37 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr2_-_71222466 | 0.37 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr4_+_69681710 | 0.36 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr1_-_13452656 | 0.36 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr9_-_125675576 | 0.36 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr5_+_98109322 | 0.36 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr15_+_101459420 | 0.36 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr5_+_138677515 | 0.36 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr7_-_22234381 | 0.36 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_-_198990166 | 0.35 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr19_+_58095501 | 0.35 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr2_+_103035102 | 0.35 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr5_+_5422778 | 0.35 |
ENST00000296564.7
|
KIAA0947
|
KIAA0947 |
| chr12_-_498620 | 0.34 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr6_+_31916733 | 0.34 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chrX_-_6453159 | 0.34 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr2_+_71357744 | 0.34 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr19_+_3933085 | 0.34 |
ENST00000168977.2
ENST00000599576.1 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr12_+_25205446 | 0.34 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr6_+_136172820 | 0.33 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr7_-_38948774 | 0.33 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr17_+_41132564 | 0.33 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr11_-_118095718 | 0.33 |
ENST00000526620.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr1_-_58716197 | 0.33 |
ENST00000371234.4
|
DAB1
|
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr5_+_138678131 | 0.33 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr4_-_74486347 | 0.32 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_+_6226782 | 0.32 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.4 | 1.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.4 | 1.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 1.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.4 | 1.4 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 1.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.5 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.2 | 0.7 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 1.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 1.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 1.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 1.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0010638 | positive regulation of organelle organization(GO:0010638) |
| 0.1 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 1.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.7 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.9 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 9.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 2.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.3 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 2.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.6 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.3 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.8 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 4.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 3.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 2.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 3.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 | 0.7 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 1.8 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 2.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.4 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 1.3 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.4 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 1.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 1.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 4.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.2 | 1.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 4.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 2.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.3 | 2.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 0.8 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.2 | 0.7 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.9 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 1.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 0.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 6.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.3 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 10.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 4.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.5 | GO:0042623 | ATPase activity, coupled(GO:0042623) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.3 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 6.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 4.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |