Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
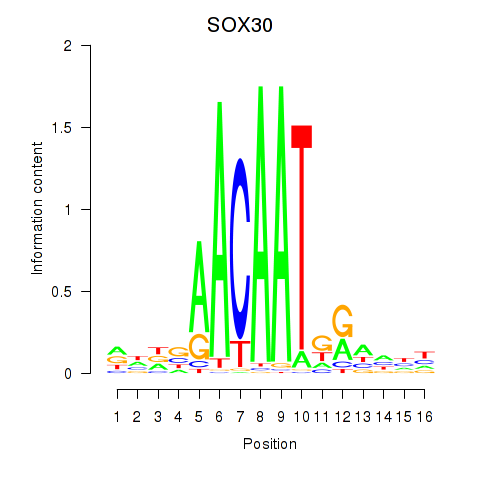
Results for SOX30
Z-value: 0.43
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079428_157079452, hg19_v2_chr5_-_157079372_157079395 | -0.24 | 2.1e-01 | Click! |
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_28270795 | 2.07 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr3_-_19988462 | 1.55 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr6_+_131958436 | 1.54 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_+_47489240 | 1.38 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr6_-_139613269 | 1.23 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr12_-_10282836 | 1.19 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_+_181429704 | 0.89 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr4_-_70725856 | 0.88 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr8_+_24151553 | 0.73 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_+_77356248 | 0.73 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr5_-_160279207 | 0.71 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr11_+_63137251 | 0.67 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr6_-_134639180 | 0.63 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_+_35415851 | 0.61 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr12_-_10282742 | 0.58 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_-_10282681 | 0.57 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_+_232573208 | 0.57 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr11_-_75236867 | 0.56 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr8_+_105352050 | 0.53 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr2_+_232573222 | 0.53 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr11_-_85430204 | 0.50 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_140235469 | 0.50 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr2_-_157198860 | 0.49 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_85430163 | 0.48 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr10_+_70480963 | 0.48 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chrX_-_117107542 | 0.41 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr8_+_24151620 | 0.40 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_-_87028478 | 0.37 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_126240442 | 0.36 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_+_36317830 | 0.32 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr5_+_170288856 | 0.29 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr11_-_85430088 | 0.28 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr22_-_30970560 | 0.25 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr22_+_46449674 | 0.23 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr1_-_85930246 | 0.23 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr13_-_41593425 | 0.22 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr7_-_92855762 | 0.22 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr5_+_139175380 | 0.22 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr15_-_52944231 | 0.21 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr6_-_49712123 | 0.21 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chrX_-_24690771 | 0.20 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chrY_+_15016013 | 0.20 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr2_+_179184955 | 0.19 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr1_+_244998602 | 0.19 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr11_-_102323740 | 0.19 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr1_+_93913713 | 0.19 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr1_-_26324534 | 0.17 |
ENST00000374284.1
ENST00000441420.1 ENST00000374282.3 |
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr5_+_140729649 | 0.17 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr3_+_140981456 | 0.17 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_+_113152978 | 0.15 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_167883327 | 0.15 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr14_+_36295504 | 0.15 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr5_-_146461027 | 0.15 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr11_-_102323489 | 0.14 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr11_-_18548426 | 0.14 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr2_-_158182410 | 0.14 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr17_-_38821373 | 0.13 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr14_-_90420862 | 0.13 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr18_+_7946941 | 0.13 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr14_-_90421028 | 0.12 |
ENST00000267544.9
ENST00000316738.7 ENST00000538485.2 ENST00000556609.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr12_+_80750134 | 0.12 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chr1_+_240255166 | 0.12 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr12_-_10251576 | 0.12 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr10_+_35416090 | 0.12 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr3_-_165555200 | 0.12 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr11_+_125365110 | 0.12 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr1_+_185014496 | 0.11 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr5_+_140787600 | 0.11 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr5_+_38148582 | 0.11 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr10_+_35416223 | 0.11 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_-_21503337 | 0.11 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr10_+_35415719 | 0.10 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr1_-_156828810 | 0.10 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr5_+_140593509 | 0.10 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr5_+_140772381 | 0.10 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr17_+_67498538 | 0.10 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_167883353 | 0.09 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr1_+_40942887 | 0.09 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr16_+_20499024 | 0.09 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr11_-_102576537 | 0.09 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr19_-_44439387 | 0.08 |
ENST00000269973.5
|
ZNF45
|
zinc finger protein 45 |
| chr4_+_54243862 | 0.08 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr12_-_10251539 | 0.08 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr4_+_71457970 | 0.07 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr2_-_37899323 | 0.07 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_-_10251603 | 0.07 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr17_+_47448102 | 0.06 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr6_+_22146851 | 0.06 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr1_+_185703513 | 0.06 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr10_+_35415978 | 0.06 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr2_+_99953816 | 0.05 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr6_-_11779174 | 0.05 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_11779014 | 0.05 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr5_-_134734901 | 0.05 |
ENST00000312469.4
ENST00000423969.2 |
H2AFY
|
H2A histone family, member Y |
| chr4_+_71458012 | 0.05 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr10_+_35484793 | 0.04 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr3_-_18466026 | 0.04 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr2_-_162931052 | 0.04 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr19_-_17375541 | 0.04 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr3_-_8686479 | 0.03 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr8_+_42195972 | 0.02 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr14_+_52456193 | 0.02 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr4_-_176733897 | 0.02 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr14_+_52456327 | 0.01 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr7_-_142176790 | 0.01 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.3 | 0.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.5 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 0.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 2.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 2.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.9 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 1.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.6 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 1.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 2.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |