Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
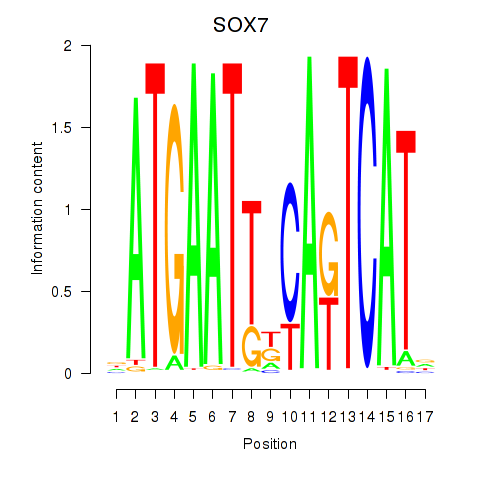
Results for SOX7
Z-value: 0.39
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg19_v2_chr8_-_10697281_10697365 | -0.06 | 7.4e-01 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_14813160 | 1.04 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr19_-_22379753 | 0.84 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr16_-_20364030 | 0.66 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr16_-_20364122 | 0.65 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr9_+_105757590 | 0.59 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr4_+_69313145 | 0.49 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr10_-_61495760 | 0.49 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr19_-_38916802 | 0.43 |
ENST00000587738.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr1_+_100598691 | 0.38 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr1_-_43751230 | 0.36 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr19_-_38916822 | 0.30 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_-_38916839 | 0.30 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr1_+_223101757 | 0.29 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr12_+_1099675 | 0.25 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr5_+_161494521 | 0.23 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr4_-_177190364 | 0.22 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr4_-_47983519 | 0.22 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr12_-_57328187 | 0.21 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr11_-_62474803 | 0.21 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr4_+_76995855 | 0.21 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr2_-_190445499 | 0.20 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr11_+_7618413 | 0.20 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_-_90049828 | 0.17 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_39734783 | 0.17 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr12_-_90049878 | 0.16 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_161494770 | 0.16 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_-_111804905 | 0.15 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr4_-_123377880 | 0.15 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr6_+_147527103 | 0.15 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr10_+_122610687 | 0.14 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr19_+_37837185 | 0.13 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr16_-_28303360 | 0.13 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr6_+_140175987 | 0.13 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr16_+_33020496 | 0.13 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr12_-_57443886 | 0.13 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr16_-_33647696 | 0.12 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr19_-_5784610 | 0.12 |
ENST00000390672.2
ENST00000419421.2 |
PRR22
|
proline rich 22 |
| chr17_-_56082455 | 0.12 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr16_+_28303804 | 0.10 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr13_-_28024681 | 0.10 |
ENST00000381116.1
ENST00000381120.3 ENST00000431572.2 |
MTIF3
|
mitochondrial translational initiation factor 3 |
| chr2_-_88285309 | 0.09 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr6_+_150690028 | 0.09 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr10_+_24738355 | 0.09 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chrY_+_26997726 | 0.09 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr2_-_166930131 | 0.08 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr2_+_87144738 | 0.08 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr1_-_26394114 | 0.08 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr2_+_3705785 | 0.06 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr5_-_95158644 | 0.06 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr11_+_3011093 | 0.06 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr6_-_86303833 | 0.05 |
ENST00000505648.1
|
SNX14
|
sorting nexin 14 |
| chr10_+_5005445 | 0.05 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr6_+_42531798 | 0.05 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_+_158901329 | 0.05 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr12_+_64173583 | 0.05 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr1_+_11724167 | 0.04 |
ENST00000376753.4
|
FBXO6
|
F-box protein 6 |
| chr8_-_19459993 | 0.04 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_102803163 | 0.04 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr5_-_95158375 | 0.04 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr6_+_80129989 | 0.03 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr6_-_3195981 | 0.02 |
ENST00000425384.2
ENST00000435043.2 |
RP1-40E16.9
|
RP1-40E16.9 |
| chr14_-_106573756 | 0.02 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr10_-_12084770 | 0.02 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr10_-_5046042 | 0.01 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr5_-_176778803 | 0.01 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr5_+_175511859 | 0.01 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr7_-_143966381 | 0.01 |
ENST00000487179.1
|
CTAGE8
|
CTAGE family, member 8 |
| chr6_-_132910877 | 0.01 |
ENST00000258034.2
|
TAAR5
|
trace amine associated receptor 5 |
| chr10_-_17243579 | 0.01 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr18_+_32820990 | 0.01 |
ENST00000601719.1
ENST00000591206.1 ENST00000330501.7 ENST00000261333.6 ENST00000355632.4 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr17_-_39623681 | 0.01 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.2 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.0 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0043605 | allantoin metabolic process(GO:0000255) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.0 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0047023 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |