Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
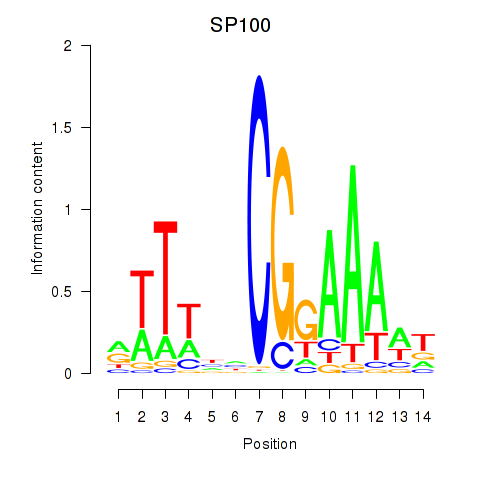
Results for SP100
Z-value: 0.83
Transcription factors associated with SP100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP100
|
ENSG00000067066.12 | SP100 nuclear antigen |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP100 | hg19_v2_chr2_+_231280954_231280981 | 0.19 | 3.2e-01 | Click! |
Activity profile of SP100 motif
Sorted Z-values of SP100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_16085314 | 4.93 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr3_-_197676740 | 3.82 |
ENST00000452735.1
ENST00000453254.1 ENST00000455191.1 |
IQCG
|
IQ motif containing G |
| chr9_+_72435709 | 2.92 |
ENST00000377197.3
ENST00000527647.1 |
C9orf135
|
chromosome 9 open reading frame 135 |
| chr6_+_88117683 | 2.37 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr6_-_33048483 | 2.36 |
ENST00000419277.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr6_+_32407619 | 2.32 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr16_+_84209539 | 2.27 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr7_-_100965011 | 2.06 |
ENST00000498704.2
ENST00000517481.1 ENST00000437644.2 ENST00000315322.4 |
RABL5
|
RAB, member RAS oncogene family-like 5 |
| chr11_-_5248294 | 1.97 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr2_+_183943464 | 1.87 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr21_-_35884573 | 1.84 |
ENST00000399286.2
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr6_+_135502408 | 1.78 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_-_52668605 | 1.72 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr12_+_111051902 | 1.69 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr2_+_39103103 | 1.57 |
ENST00000340556.6
ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2
|
MORN repeat containing 2 |
| chr11_+_73661364 | 1.52 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr12_-_58329819 | 1.51 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr2_-_178417742 | 1.48 |
ENST00000408939.3
|
TTC30B
|
tetratricopeptide repeat domain 30B |
| chr20_-_45318230 | 1.46 |
ENST00000372114.3
|
TP53RK
|
TP53 regulating kinase |
| chr6_+_32709119 | 1.45 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr6_+_135502466 | 1.38 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_+_47489240 | 1.36 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr1_+_63989004 | 1.33 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr17_+_7761013 | 1.27 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr19_+_4639514 | 1.26 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr17_+_7761301 | 1.21 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr5_+_35617940 | 1.19 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr5_+_140514782 | 1.09 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr8_+_94767072 | 1.07 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr3_+_113666748 | 1.01 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr10_-_46167722 | 0.94 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chrX_-_77395186 | 0.94 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr5_-_41261540 | 0.94 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr8_+_75262612 | 0.90 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr15_+_45879534 | 0.90 |
ENST00000564080.1
ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4
BLOC1S6
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr10_-_29084886 | 0.88 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr4_+_41540160 | 0.87 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_10764509 | 0.86 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr15_+_66797627 | 0.85 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr3_-_122134882 | 0.84 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr10_+_127661942 | 0.83 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr6_+_42847649 | 0.80 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr16_+_16484691 | 0.78 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr21_+_40752170 | 0.78 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr12_+_122356488 | 0.78 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr12_-_25801478 | 0.77 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr15_-_45670924 | 0.77 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr7_-_120498357 | 0.74 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr12_+_123459127 | 0.74 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr2_-_28113965 | 0.74 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr12_-_42877726 | 0.74 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr15_-_93632421 | 0.72 |
ENST00000329082.7
|
RGMA
|
repulsive guidance molecule family member a |
| chr12_+_120884222 | 0.71 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr4_+_128802016 | 0.70 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr2_+_183989083 | 0.69 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr13_+_25254545 | 0.69 |
ENST00000218548.6
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr11_-_77790865 | 0.69 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr1_-_234614849 | 0.68 |
ENST00000040877.1
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1 |
| chr18_-_52989525 | 0.68 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr11_-_77791156 | 0.68 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr21_+_40759684 | 0.68 |
ENST00000380708.1
|
WRB
|
tryptophan rich basic protein |
| chr11_-_5255696 | 0.66 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr4_+_89299994 | 0.66 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_-_45883558 | 0.66 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr15_+_66797455 | 0.66 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr2_+_183989157 | 0.65 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr8_+_110552831 | 0.64 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr17_+_17876127 | 0.64 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr6_-_109804412 | 0.62 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chrX_-_153707246 | 0.61 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3 |
| chr11_-_17410869 | 0.60 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr5_+_148651409 | 0.60 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr3_+_130745688 | 0.60 |
ENST00000510769.1
ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11
|
NIMA-related kinase 11 |
| chr6_+_29691056 | 0.59 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_-_160001737 | 0.59 |
ENST00000368090.2
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr15_-_93616340 | 0.59 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr3_-_11685345 | 0.58 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_-_119256311 | 0.58 |
ENST00000316316.6
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr1_+_227751231 | 0.58 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr10_+_38299546 | 0.58 |
ENST00000374618.3
ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A
|
zinc finger protein 33A |
| chr16_+_66914264 | 0.57 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr14_-_24911448 | 0.57 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr7_-_117513540 | 0.56 |
ENST00000160373.3
|
CTTNBP2
|
cortactin binding protein 2 |
| chr18_-_52989217 | 0.56 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_-_110011718 | 0.56 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chrX_-_153707545 | 0.56 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr8_+_75262629 | 0.56 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr6_+_135502501 | 0.55 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_-_109203997 | 0.55 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr1_+_15272271 | 0.54 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chrX_-_10588459 | 0.53 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr7_-_32529973 | 0.53 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_+_46859933 | 0.53 |
ENST00000243167.8
|
FAAH
|
fatty acid amide hydrolase |
| chr13_+_114462193 | 0.52 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr6_+_29691198 | 0.52 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr3_+_62304648 | 0.51 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr17_+_16284604 | 0.51 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr12_+_104682496 | 0.51 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr1_+_13521973 | 0.51 |
ENST00000327795.5
|
PRAMEF21
|
PRAME family member 21 |
| chr3_-_43147549 | 0.50 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr14_-_75530693 | 0.50 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_+_2487800 | 0.50 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr12_-_110841462 | 0.50 |
ENST00000455511.3
ENST00000450008.2 |
ANAPC7
|
anaphase promoting complex subunit 7 |
| chr8_+_110552337 | 0.49 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr1_+_231473743 | 0.49 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr11_-_76381781 | 0.49 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr17_-_62502022 | 0.48 |
ENST00000578804.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr9_-_126030817 | 0.48 |
ENST00000348403.5
ENST00000447404.2 ENST00000360998.3 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr6_-_167571817 | 0.47 |
ENST00000366834.1
|
GPR31
|
G protein-coupled receptor 31 |
| chrX_-_10588595 | 0.47 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr9_-_135754164 | 0.47 |
ENST00000298545.3
|
AK8
|
adenylate kinase 8 |
| chr14_-_24912047 | 0.47 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr2_-_89399845 | 0.46 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr19_+_32896646 | 0.46 |
ENST00000392250.2
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr14_-_102605983 | 0.46 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr16_+_84209738 | 0.45 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr6_-_13486369 | 0.45 |
ENST00000558378.1
|
AL583828.1
|
AL583828.1 |
| chr17_-_4544960 | 0.45 |
ENST00000293761.3
|
ALOX15
|
arachidonate 15-lipoxygenase |
| chr14_-_24911868 | 0.45 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr5_-_76383133 | 0.45 |
ENST00000255198.2
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr15_+_72410629 | 0.44 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr19_+_56989609 | 0.44 |
ENST00000601875.1
|
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr20_+_42219559 | 0.44 |
ENST00000373030.3
ENST00000373039.4 |
IFT52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr10_+_91174314 | 0.44 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr13_-_31736132 | 0.44 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_-_131949200 | 0.43 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr9_-_135230336 | 0.42 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr17_-_62502399 | 0.42 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr16_-_28303360 | 0.42 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr14_+_22446680 | 0.42 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr13_-_31736027 | 0.41 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr9_-_97402531 | 0.41 |
ENST00000415431.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr2_+_179149636 | 0.41 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr7_-_156685841 | 0.40 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr1_-_39339777 | 0.40 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr10_-_61495760 | 0.39 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr14_+_105957402 | 0.39 |
ENST00000421892.1
ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80
|
chromosome 14 open reading frame 80 |
| chr19_+_54024251 | 0.39 |
ENST00000253144.9
|
ZNF331
|
zinc finger protein 331 |
| chr14_-_24911971 | 0.39 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr3_-_15106747 | 0.38 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr6_+_32812568 | 0.38 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr10_+_60272814 | 0.38 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr9_-_97402413 | 0.38 |
ENST00000414122.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr10_+_35416223 | 0.37 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chrX_+_144908928 | 0.37 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr3_+_119187785 | 0.37 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr12_+_4758264 | 0.37 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr4_+_159690218 | 0.37 |
ENST00000264433.6
|
FNIP2
|
folliculin interacting protein 2 |
| chrX_+_77154935 | 0.36 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr6_+_41888926 | 0.36 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr10_+_51187938 | 0.36 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr3_-_186288097 | 0.36 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr6_+_10585979 | 0.36 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr16_-_4664860 | 0.36 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr2_+_38893208 | 0.36 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr4_+_40058411 | 0.36 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr2_-_70418032 | 0.35 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr11_-_76381029 | 0.35 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr16_-_30441293 | 0.35 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr2_+_39005336 | 0.35 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr4_+_89299885 | 0.35 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr16_-_68057770 | 0.35 |
ENST00000332395.5
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| chrX_+_36246735 | 0.35 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr22_+_32455111 | 0.35 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr14_-_102552659 | 0.35 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr8_+_39442097 | 0.35 |
ENST00000265707.5
ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18
|
ADAM metallopeptidase domain 18 |
| chr14_-_21490417 | 0.35 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chrX_+_11129388 | 0.34 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr2_+_189156586 | 0.34 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_-_111334678 | 0.34 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr14_-_21490653 | 0.34 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr6_+_158957431 | 0.34 |
ENST00000367090.3
|
TMEM181
|
transmembrane protein 181 |
| chr15_+_93749295 | 0.34 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr9_-_32526184 | 0.33 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chrX_+_108780347 | 0.33 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr5_+_98104978 | 0.33 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chr17_+_21030260 | 0.33 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr4_+_69962185 | 0.32 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr2_-_152118352 | 0.32 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr19_+_52901094 | 0.32 |
ENST00000391788.2
ENST00000436397.1 ENST00000391787.2 ENST00000360465.3 ENST00000494167.2 ENST00000493272.1 |
ZNF528
|
zinc finger protein 528 |
| chr14_-_20774092 | 0.32 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr9_-_99775862 | 0.32 |
ENST00000602917.1
ENST00000375223.4 |
HIATL2
|
hippocampus abundant transcript-like 2 |
| chr9_-_139343294 | 0.32 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr11_-_31531121 | 0.31 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr6_-_131949305 | 0.31 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr15_+_90777424 | 0.30 |
ENST00000561433.1
ENST00000559204.1 ENST00000558291.1 |
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr11_+_7626950 | 0.30 |
ENST00000530181.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr13_-_31736478 | 0.30 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr14_-_21490590 | 0.30 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chrX_+_70586140 | 0.30 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr1_-_167883327 | 0.30 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr5_+_40841276 | 0.30 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr7_+_12610307 | 0.29 |
ENST00000297029.5
|
SCIN
|
scinderin |
| chr2_-_55920952 | 0.29 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr19_+_56159362 | 0.29 |
ENST00000593069.1
ENST00000308964.3 |
CCDC106
|
coiled-coil domain containing 106 |
| chr6_-_41888814 | 0.29 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr19_+_21688366 | 0.28 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr22_-_20307532 | 0.28 |
ENST00000405465.3
ENST00000248879.3 |
DGCR6L
|
DiGeorge syndrome critical region gene 6-like |
| chrX_-_20159934 | 0.28 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr9_-_33264557 | 0.28 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of SP100
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.2 | 4.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.6 | 2.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.5 | 2.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.5 | 2.7 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.4 | 3.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 1.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.7 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 0.7 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 0.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 0.7 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 0.2 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.2 | 1.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 | 0.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.8 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 0.6 | GO:0006186 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.2 | 0.7 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.4 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.7 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 1.2 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.7 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 2.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 1.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.3 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.6 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.5 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 1.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.4 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.0 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.4 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.9 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.3 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.2 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 1.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 2.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 3.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 1.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 1.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.4 | GO:0031334 | positive regulation of protein complex assembly(GO:0031334) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 1.0 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.5 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 1.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.3 | GO:0070646 | protein deubiquitination(GO:0016579) protein modification by small protein removal(GO:0070646) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0002177 | manchette(GO:0002177) |
| 0.5 | 2.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 6.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 4.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 0.8 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 0.7 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 0.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 2.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0042612 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.1 | 1.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 4.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.6 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 2.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 3.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 0.5 | GO:0004040 | amidase activity(GO:0004040) |
| 0.2 | 0.7 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 0.7 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.5 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.2 | 1.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 1.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.6 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 3.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 4.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 2.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0019784 | SUMO-specific protease activity(GO:0016929) NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 1.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |