Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
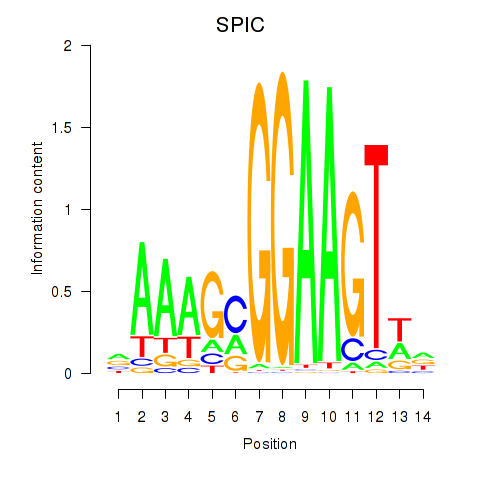
Results for SPIC
Z-value: 0.93
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIC | hg19_v2_chr12_+_101869096_101869199 | 0.14 | 4.6e-01 | Click! |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_8201091 | 5.32 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr19_-_51471362 | 4.04 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr19_-_51471381 | 3.96 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr12_+_15699286 | 3.37 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr2_-_113594279 | 3.00 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr12_+_8975061 | 2.90 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr3_-_111314230 | 2.70 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr18_-_33077556 | 2.61 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr4_-_80994471 | 2.57 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr2_+_233925064 | 2.43 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_-_15344243 | 2.35 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr19_+_35645618 | 2.32 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr4_-_57547454 | 2.32 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr2_+_113885138 | 2.31 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr19_-_51456344 | 2.28 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr19_-_51456321 | 2.26 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr15_-_80263506 | 2.19 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr4_-_80994619 | 2.08 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr19_-_51456198 | 2.06 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr1_-_43855444 | 1.96 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr4_-_111119804 | 1.93 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_+_120839412 | 1.92 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr12_+_1800179 | 1.92 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr19_+_35645817 | 1.89 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr1_+_151030234 | 1.78 |
ENST00000368921.3
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 |
| chr22_-_37880543 | 1.72 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_206138884 | 1.67 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr1_-_143913143 | 1.64 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr1_+_206138457 | 1.61 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr7_+_74188309 | 1.57 |
ENST00000289473.4
ENST00000433458.1 |
NCF1
|
neutrophil cytosolic factor 1 |
| chr1_+_203651937 | 1.56 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr1_+_120839005 | 1.56 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr19_-_42927251 | 1.55 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr1_-_27816556 | 1.52 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr10_+_102106829 | 1.52 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr16_+_23652773 | 1.49 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr6_+_106534192 | 1.48 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr2_-_69098566 | 1.48 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr16_+_30194916 | 1.46 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr18_-_33077942 | 1.44 |
ENST00000334598.7
|
INO80C
|
INO80 complex subunit C |
| chr2_-_238323007 | 1.44 |
ENST00000295550.4
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr5_+_96211643 | 1.41 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr1_+_192127578 | 1.40 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr11_+_68671310 | 1.39 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr12_+_120105558 | 1.38 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr5_-_39270725 | 1.37 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr10_-_98031265 | 1.35 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr9_-_95640218 | 1.34 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr15_+_63354769 | 1.33 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_+_47324424 | 1.32 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr1_-_6420737 | 1.28 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr11_-_64647144 | 1.26 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr8_-_144655141 | 1.26 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chrX_-_107018969 | 1.25 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr16_+_11439286 | 1.23 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr11_-_64851496 | 1.22 |
ENST00000404147.3
ENST00000275517.3 |
CDCA5
|
cell division cycle associated 5 |
| chr1_-_154946825 | 1.21 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_+_130613226 | 1.20 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_-_98031310 | 1.18 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr10_-_121302195 | 1.17 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr16_-_88851618 | 1.15 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr17_+_1959369 | 1.14 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr5_+_33936491 | 1.13 |
ENST00000330120.3
|
RXFP3
|
relaxin/insulin-like family peptide receptor 3 |
| chr15_+_90744533 | 1.13 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr11_-_1643368 | 1.11 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr10_-_75401500 | 1.11 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr1_+_24645807 | 1.11 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr16_-_20702578 | 1.10 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr17_-_37009882 | 1.08 |
ENST00000378096.3
ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23
|
ribosomal protein L23 |
| chr9_+_116037922 | 1.07 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr20_-_54967187 | 1.05 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr3_+_130569592 | 1.04 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_169337172 | 1.04 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr2_+_48667983 | 1.04 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr1_-_150780757 | 1.04 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr12_-_51785182 | 1.04 |
ENST00000356317.3
ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr1_+_110091189 | 1.02 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr4_-_159094194 | 1.00 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr11_+_70244510 | 0.99 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr1_-_24127256 | 0.98 |
ENST00000418277.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr1_+_24645865 | 0.98 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_+_121796697 | 0.98 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr20_+_814377 | 0.97 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr17_-_43487741 | 0.97 |
ENST00000455881.1
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr10_+_112257596 | 0.97 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr1_+_150245099 | 0.96 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr4_-_80994210 | 0.95 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_+_150245177 | 0.94 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr19_+_19496728 | 0.94 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr2_+_48667898 | 0.94 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr1_-_47069886 | 0.94 |
ENST00000371946.4
ENST00000371945.4 ENST00000428112.2 ENST00000529170.1 |
MKNK1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr1_-_52499443 | 0.94 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr15_-_33360342 | 0.93 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr1_+_11994715 | 0.92 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chrX_-_51239425 | 0.91 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr11_+_133938820 | 0.91 |
ENST00000299106.4
ENST00000529443.2 |
JAM3
|
junctional adhesion molecule 3 |
| chr16_+_30205225 | 0.91 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr12_-_49351228 | 0.90 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr9_-_134145880 | 0.90 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr12_-_54778471 | 0.90 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr16_-_29465668 | 0.90 |
ENST00000569622.1
|
RP11-345J4.5
|
BolA-like protein 2 |
| chr17_+_48450575 | 0.90 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr12_-_7596735 | 0.89 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr16_+_29465822 | 0.89 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr1_-_47069955 | 0.89 |
ENST00000341183.5
ENST00000496619.1 |
MKNK1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr15_-_34610962 | 0.88 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_+_154393260 | 0.88 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chrX_+_24072833 | 0.87 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr2_+_37458928 | 0.86 |
ENST00000439218.1
ENST00000432075.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr12_+_53693466 | 0.86 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr3_-_28390120 | 0.85 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr10_-_75676400 | 0.84 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr1_-_180991978 | 0.84 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr1_+_22979474 | 0.83 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr3_+_52321827 | 0.82 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr4_-_99064387 | 0.82 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr18_-_33077868 | 0.82 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr11_-_102668879 | 0.81 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr14_-_69445968 | 0.81 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr1_+_212208919 | 0.80 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr12_-_113841678 | 0.80 |
ENST00000552280.1
ENST00000257549.4 |
SDS
|
serine dehydratase |
| chr12_-_77272765 | 0.78 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr19_+_41256764 | 0.77 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_-_32145861 | 0.77 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr14_-_69446034 | 0.76 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr12_+_10658489 | 0.76 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr14_-_69445793 | 0.76 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chrX_+_51075658 | 0.76 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr13_+_31309645 | 0.75 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr19_+_38826477 | 0.75 |
ENST00000409410.2
ENST00000215069.4 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chrX_+_30233668 | 0.75 |
ENST00000378988.4
|
MAGEB2
|
melanoma antigen family B, 2 |
| chr4_-_99578776 | 0.74 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chrX_+_100075368 | 0.74 |
ENST00000415585.2
ENST00000372972.2 ENST00000413437.1 |
CSTF2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
| chr20_+_3869423 | 0.74 |
ENST00000497424.1
|
PANK2
|
pantothenate kinase 2 |
| chr14_+_67291158 | 0.74 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr22_+_37309662 | 0.74 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr5_+_110074685 | 0.73 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr4_-_74853897 | 0.73 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr3_-_179169330 | 0.73 |
ENST00000232564.3
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr5_+_115177178 | 0.72 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_-_28390298 | 0.72 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_138665969 | 0.72 |
ENST00000330315.3
|
FOXL2
|
forkhead box L2 |
| chr1_-_109618566 | 0.72 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr8_-_8243968 | 0.71 |
ENST00000520004.1
|
SGK223
|
Tyrosine-protein kinase SgK223 |
| chr15_-_67546963 | 0.71 |
ENST00000561452.1
ENST00000261880.5 |
AAGAB
|
alpha- and gamma-adaptin binding protein |
| chr11_+_61891445 | 0.71 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr3_-_156272924 | 0.70 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr8_-_42623747 | 0.70 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr7_-_80551671 | 0.70 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chrX_-_153637612 | 0.69 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr19_+_45417504 | 0.69 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr11_+_72983246 | 0.69 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr6_-_136571400 | 0.69 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr13_+_41363581 | 0.68 |
ENST00000338625.4
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
| chr12_-_109915098 | 0.68 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr2_-_70780572 | 0.68 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr18_-_61089665 | 0.68 |
ENST00000238497.5
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr7_-_99698338 | 0.68 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_-_24126892 | 0.67 |
ENST00000374497.3
ENST00000425913.1 |
GALE
|
UDP-galactose-4-epimerase |
| chr11_-_67120974 | 0.67 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr4_-_89152474 | 0.67 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_+_22979676 | 0.67 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr3_-_196014520 | 0.66 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr6_-_27114577 | 0.66 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr1_-_94147385 | 0.66 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr17_+_42925270 | 0.66 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr4_+_57333756 | 0.66 |
ENST00000510663.1
ENST00000504757.1 |
SRP72
|
signal recognition particle 72kDa |
| chr3_-_50375657 | 0.65 |
ENST00000395126.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr8_-_42623924 | 0.65 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr17_+_7482785 | 0.65 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr1_+_33938236 | 0.64 |
ENST00000361328.3
ENST00000373413.2 |
ZSCAN20
|
zinc finger and SCAN domain containing 20 |
| chr13_+_49280951 | 0.64 |
ENST00000282018.3
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr1_-_9129735 | 0.64 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr20_-_1309809 | 0.64 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr18_-_34408802 | 0.63 |
ENST00000590842.1
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr3_-_28390415 | 0.63 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr11_-_85565906 | 0.62 |
ENST00000544076.1
|
AP000974.1
|
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
| chr11_+_65408273 | 0.62 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr5_+_96079240 | 0.62 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr4_-_99578789 | 0.61 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr3_+_148847371 | 0.61 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr14_+_75469606 | 0.61 |
ENST00000266126.5
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr16_+_15737124 | 0.61 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr19_+_1752372 | 0.61 |
ENST00000382349.4
|
ONECUT3
|
one cut homeobox 3 |
| chr9_-_36276966 | 0.60 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr12_-_95044309 | 0.60 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_-_72206034 | 0.60 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr3_-_156272872 | 0.60 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr20_-_44540686 | 0.59 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr5_+_74062806 | 0.59 |
ENST00000296802.5
|
NSA2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr1_-_161519682 | 0.59 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr2_-_101925055 | 0.59 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr5_-_140013275 | 0.59 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr16_+_23652700 | 0.59 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr20_-_48532046 | 0.58 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr1_-_175161890 | 0.58 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr17_-_5138099 | 0.58 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr3_-_149095652 | 0.58 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr17_-_76123101 | 0.58 |
ENST00000392467.3
|
TMC6
|
transmembrane channel-like 6 |
| chr19_+_38826415 | 0.58 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.1 | 3.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 1.1 | 6.6 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.0 | 3.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.8 | 2.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.5 | 1.6 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.5 | 2.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 1.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 0.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.5 | 2.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.4 | 1.2 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 0.4 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.4 | 1.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.4 | 8.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 1.1 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.4 | 1.1 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.3 | 1.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.3 | 1.3 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 1.9 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.3 | 1.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.9 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 1.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 0.9 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.5 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.3 | 0.9 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.3 | 0.9 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.1 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.3 | 1.7 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 1.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 2.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 1.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.3 | 2.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.7 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.7 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 0.7 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 0.9 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 0.4 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 1.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 1.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.2 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.2 | 0.8 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.6 | GO:0043473 | pigmentation(GO:0043473) |
| 0.2 | 0.8 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 | 0.2 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.6 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 0.9 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 0.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.5 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 | 0.7 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 | 2.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 0.9 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 0.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.4 | GO:0043318 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.1 | 0.6 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.6 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.7 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.7 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.0 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.7 | GO:0033686 | oocyte growth(GO:0001555) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 1.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.8 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 2.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.1 | 0.4 | GO:0100009 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.4 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.7 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 1.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 1.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 0.5 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.7 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.7 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.7 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.8 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 1.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 2.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.4 | GO:2001191 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 0.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.8 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 1.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.4 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.5 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.1 | 0.9 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.1 | 2.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.2 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.3 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.3 | GO:0032680 | tumor necrosis factor production(GO:0032640) regulation of tumor necrosis factor production(GO:0032680) regulation of tumor necrosis factor superfamily cytokine production(GO:1903555) |
| 0.1 | 2.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 3.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:1905154 | negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 1.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.4 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.7 | GO:0015886 | heme transport(GO:0015886) xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.8 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 0.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.5 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.5 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 1.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.3 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:1903978 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.2 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:2000416 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 2.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.0 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.5 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.6 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.0 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.5 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 1.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 1.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.5 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.7 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 1.0 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.3 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 2.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 1.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.4 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 1.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 1.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 3.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 1.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 3.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 1.5 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.3 | 1.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.3 | 1.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 1.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 1.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 0.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 2.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 4.9 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.6 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 1.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 2.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 1.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) alveolar lamellar body(GO:0097208) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0031390 | DNA replication factor C complex(GO:0005663) Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 4.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 4.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.6 | 2.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.5 | 1.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.5 | 1.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.4 | 1.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.7 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 2.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 0.9 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 1.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 0.9 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.3 | 1.7 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.3 | 1.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 2.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 0.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 0.8 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.2 | 0.7 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.7 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.2 | 1.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 1.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 0.6 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 0.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 2.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 0.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 0.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 3.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 0.5 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 1.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.5 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.2 | 3.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 1.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.7 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.3 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 1.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 3.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 2.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.3 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.8 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 3.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 4.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 13.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0001032 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0070990 | snRNP binding(GO:0070990) U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0035242 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 8.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 5.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 2.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 8.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 7.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.4 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 5.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |