Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
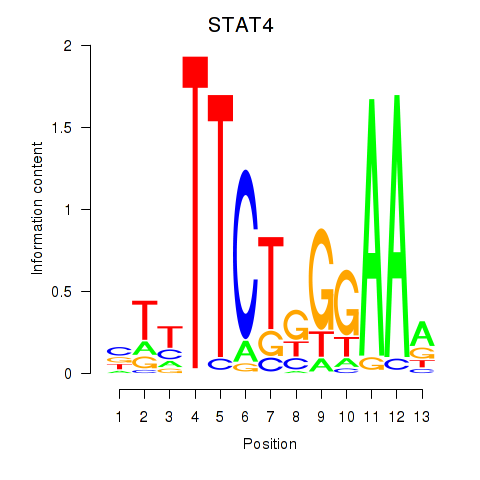
Results for STAT4
Z-value: 0.68
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg19_v2_chr2_-_192016316_192016325 | -0.53 | 2.6e-03 | Click! |
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_161337662 | 4.21 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr6_+_32407619 | 4.15 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr14_-_21270561 | 3.02 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr1_-_109655377 | 2.92 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_-_109655355 | 2.91 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr6_+_32821924 | 1.90 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_+_140227048 | 1.83 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr5_-_121413974 | 1.76 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr5_+_140261703 | 1.74 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr16_+_19467772 | 1.70 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr3_-_148939835 | 1.65 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_+_58283814 | 1.44 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr5_-_16509101 | 1.32 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr12_+_113587558 | 1.22 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr19_+_35168567 | 1.07 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
| chr19_+_35225060 | 1.04 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr5_-_111092873 | 1.02 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr6_-_110011704 | 1.01 |
ENST00000448084.2
|
AK9
|
adenylate kinase 9 |
| chr10_-_116444371 | 0.98 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr6_+_32811861 | 0.97 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr7_+_117120017 | 0.97 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr5_-_111093406 | 0.95 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr3_-_114866084 | 0.94 |
ENST00000357258.3
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_110011718 | 0.94 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr19_+_35168633 | 0.93 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr5_-_111092930 | 0.91 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr22_-_38349552 | 0.86 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr5_-_111093167 | 0.86 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr12_-_15082050 | 0.85 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chrX_+_144908928 | 0.78 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr5_-_111093340 | 0.70 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr19_-_50529193 | 0.70 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr20_-_21494654 | 0.69 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr11_+_6411670 | 0.69 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr11_-_118436707 | 0.68 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr19_+_44669280 | 0.68 |
ENST00000590089.1
ENST00000454662.2 |
ZNF226
|
zinc finger protein 226 |
| chr11_+_117070904 | 0.68 |
ENST00000529792.1
|
TAGLN
|
transgelin |
| chr16_-_67260691 | 0.65 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr5_-_111093081 | 0.65 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr3_+_171561127 | 0.62 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr9_+_100069933 | 0.60 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr14_-_21490653 | 0.57 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21490417 | 0.56 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr16_+_78056412 | 0.56 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chrX_-_137793826 | 0.54 |
ENST00000315930.6
|
FGF13
|
fibroblast growth factor 13 |
| chr14_-_21490958 | 0.53 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr1_+_202431859 | 0.53 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_+_123986069 | 0.53 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr12_+_58138800 | 0.51 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr14_-_21490590 | 0.50 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chr7_-_92855762 | 0.49 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr5_+_73109339 | 0.49 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr12_+_58138664 | 0.48 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr10_-_104179682 | 0.46 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr10_+_102790980 | 0.46 |
ENST00000393459.1
ENST00000224807.5 |
SFXN3
|
sideroflexin 3 |
| chr18_+_6729698 | 0.45 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr13_+_97928395 | 0.44 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr21_+_38593701 | 0.43 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr6_-_32811771 | 0.42 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr1_-_120935894 | 0.42 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr19_-_51875894 | 0.42 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr5_-_114632307 | 0.40 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr9_-_116163400 | 0.37 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr2_-_160473114 | 0.36 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_-_156534754 | 0.36 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr2_-_79386786 | 0.36 |
ENST00000393878.1
ENST00000305165.2 ENST00000409839.3 |
REG3A
|
regenerating islet-derived 3 alpha |
| chr22_-_31536480 | 0.35 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chrX_-_46187069 | 0.35 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr19_+_44331493 | 0.34 |
ENST00000588797.1
|
ZNF283
|
zinc finger protein 283 |
| chr2_-_160472952 | 0.34 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_+_9299895 | 0.33 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr1_-_144994909 | 0.33 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chrX_+_135570046 | 0.32 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr16_+_4674814 | 0.32 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr19_+_39881951 | 0.31 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr7_+_117120106 | 0.31 |
ENST00000426809.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr2_-_29297127 | 0.30 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr19_+_10197463 | 0.29 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr4_-_187476721 | 0.29 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr3_-_183735651 | 0.29 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_+_44331444 | 0.29 |
ENST00000324461.7
|
ZNF283
|
zinc finger protein 283 |
| chr11_+_118175596 | 0.28 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr13_-_41240717 | 0.27 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr16_+_4674787 | 0.26 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr3_-_187455680 | 0.26 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr16_+_67261008 | 0.25 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr14_-_20881579 | 0.25 |
ENST00000556935.1
ENST00000262715.5 ENST00000556549.1 |
TEP1
|
telomerase-associated protein 1 |
| chr16_+_66600294 | 0.25 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr17_-_77924627 | 0.25 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr6_+_46761118 | 0.24 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr11_-_10590238 | 0.23 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_-_27468842 | 0.23 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr6_+_52051171 | 0.22 |
ENST00000340057.1
|
IL17A
|
interleukin 17A |
| chr4_+_130017268 | 0.22 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr6_-_49712123 | 0.22 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr19_+_50528971 | 0.22 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr11_+_118175132 | 0.21 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chrX_-_70474910 | 0.21 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr15_-_52944231 | 0.20 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr1_+_40997233 | 0.20 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr15_+_65843130 | 0.20 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr6_-_136610911 | 0.19 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr15_+_45021183 | 0.19 |
ENST00000559390.1
|
TRIM69
|
tripartite motif containing 69 |
| chr16_-_67260901 | 0.19 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr13_+_100153665 | 0.19 |
ENST00000376387.4
|
TM9SF2
|
transmembrane 9 superfamily member 2 |
| chr13_-_45915221 | 0.19 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr1_-_231005310 | 0.19 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr22_+_24551765 | 0.19 |
ENST00000337989.7
|
CABIN1
|
calcineurin binding protein 1 |
| chrX_-_65253506 | 0.19 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr4_-_140005443 | 0.19 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chrX_-_154563889 | 0.18 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr9_-_116837249 | 0.18 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr12_-_49245936 | 0.18 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr1_+_161551101 | 0.18 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr11_-_119999539 | 0.18 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr6_+_134210243 | 0.18 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr20_+_5731083 | 0.18 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr12_-_55367361 | 0.17 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr14_+_65381079 | 0.17 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr14_+_56127989 | 0.17 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr19_+_50529212 | 0.16 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr12_-_11175219 | 0.16 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr3_-_52864680 | 0.16 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr11_-_119999611 | 0.16 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chr5_+_140753444 | 0.16 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr2_-_202563414 | 0.16 |
ENST00000409474.3
ENST00000315506.7 ENST00000359962.5 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr3_+_122296465 | 0.15 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr17_+_41132564 | 0.15 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr9_+_72658490 | 0.15 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr17_-_58603568 | 0.14 |
ENST00000083182.3
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr5_+_40909354 | 0.14 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr19_+_37709076 | 0.14 |
ENST00000590503.1
ENST00000589413.1 |
ZNF383
|
zinc finger protein 383 |
| chr20_-_18774614 | 0.14 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr2_+_234826016 | 0.14 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_+_57998821 | 0.14 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr14_-_25103472 | 0.13 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_+_45429998 | 0.13 |
ENST00000265537.3
ENST00000415258.1 ENST00000431023.1 ENST00000414984.1 |
LARS2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr6_-_25874440 | 0.13 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr2_-_202562774 | 0.13 |
ENST00000396886.3
ENST00000409143.1 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr1_-_162346657 | 0.13 |
ENST00000367935.5
|
C1orf111
|
chromosome 1 open reading frame 111 |
| chr1_+_149754227 | 0.13 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr16_-_46782221 | 0.12 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr15_+_57998923 | 0.11 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr1_-_8000872 | 0.11 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_+_20848282 | 0.11 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr20_+_8112824 | 0.10 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr2_-_177502254 | 0.10 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr17_-_34207295 | 0.10 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr10_+_17794251 | 0.10 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr3_+_122399444 | 0.10 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr7_-_82792215 | 0.10 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr12_+_20848377 | 0.10 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr15_-_60690163 | 0.10 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr14_-_21924209 | 0.10 |
ENST00000557364.1
|
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr8_-_123139423 | 0.09 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chrX_+_149861836 | 0.09 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr16_+_24549014 | 0.09 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr10_+_104404644 | 0.09 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr6_-_116833500 | 0.09 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr19_-_56904799 | 0.09 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr6_+_25754927 | 0.09 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr19_-_7812397 | 0.08 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr10_+_6625733 | 0.08 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr5_+_32585605 | 0.08 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr14_+_69658480 | 0.08 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chrX_+_24711997 | 0.07 |
ENST00000379068.3
ENST00000379059.3 |
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr1_-_161519682 | 0.07 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr14_-_81408063 | 0.07 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr4_-_140477353 | 0.07 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr13_-_26795840 | 0.07 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_+_42131011 | 0.07 |
ENST00000458483.1
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic) |
| chr1_+_50571949 | 0.07 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_-_185597619 | 0.07 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr4_-_46126093 | 0.07 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr19_-_44405623 | 0.06 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr17_-_27038063 | 0.06 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr12_+_70636765 | 0.06 |
ENST00000552231.1
ENST00000229195.3 ENST00000547780.1 ENST00000418359.3 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr3_+_77088989 | 0.06 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr20_+_39657454 | 0.06 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr6_+_31082603 | 0.06 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr8_-_114449112 | 0.06 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr19_-_7812446 | 0.05 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr17_-_5323480 | 0.05 |
ENST00000573584.1
|
NUP88
|
nucleoporin 88kDa |
| chr8_-_27469196 | 0.05 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr3_+_155838337 | 0.04 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr14_+_24630465 | 0.04 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr19_-_54663473 | 0.04 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr12_-_11139511 | 0.04 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr19_-_39881777 | 0.04 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr6_+_108977520 | 0.03 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr2_-_37193606 | 0.03 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chrX_-_107334790 | 0.03 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr1_+_179699318 | 0.03 |
ENST00000423879.1
|
RP11-12M5.1
|
RP11-12M5.1 |
| chr1_-_144994840 | 0.03 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_-_84686552 | 0.03 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr9_-_27297126 | 0.03 |
ENST00000380031.1
ENST00000537675.1 ENST00000380032.3 |
EQTN
|
equatorin, sperm acrosome associated |
| chr19_-_44405941 | 0.03 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr12_+_20848486 | 0.03 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chrX_+_100646190 | 0.03 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr19_+_44669221 | 0.02 |
ENST00000590578.1
ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226
|
zinc finger protein 226 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.7 | 2.0 | GO:0061567 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.4 | 1.3 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 0.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 0.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 1.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.7 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.2 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.1 | 1.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 3.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) |
| 0.0 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 2.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:0043317 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 4.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.7 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.4 | 1.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.4 | 1.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 4.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 2.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 1.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 2.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |